Gene
KWMTBOMO00334
Pre Gene Modal
BGIBMGA000699
Annotation
PREDICTED:_amyloid_beta_A4_precursor_protein-binding_family_B_member_1-interacting_protein_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.265
Sequence
CDS
ATGGCGTTGGTAGAGGAAGTCGGTGCGGGCTCCACCGCTGACGATCCCGAAGCTCTTCTGAACGAGTGGCTTGGCGAACTGACCGTTCTTACTGCGGGTCTTAATTCAACCGGCGATGGTTCATTAGCTCTGAGGCCTCTTGAAATAGTCGCTCCACGCATCGACACCTATCGGTTCTCGATGGCAAATCTCGAAGAAACGCAAGACGCTGATCTTGACGCCATTCTGGGCGAACTATGCGCTCTCGATTCCGAATATGACGAAGAATTATCTAGAGTGTCATCAGGCTTTGCATCCGGATCAAAAGACAGGGTACCGAGAACCACTGAGCCCTGTACCGTGCGTCAGGAGAAGGACAACAGCGATGCGGCTTCGGGGATCACGCGCACCGAGTCCCCGGACAATGACTCCGCTTTCAGTGACACAGTATCCATGCTTTCGAGCGAATCTTCGGCTTCTAGTAGCGCGAGTGCGAAATGCAAAGCCTTAAAATTGAATTTACATCAAAATCAAAAGGACGCGAGCTTCCAACAAAAGGCGGACAAGATTAAATTAGCATTAGAGCGCATGCGAGAAGCAAATATCAAGAAACTGTTCATAAAAGCGTTTTCAATGGATGGATCTTCGAAGAGTTTATTGGTTGATGAGAAAATGACTTGTGGATATGTGACGCGTTTGCTGGCGGACAAAAACCACGTTAATATGGAACCGAAGTGGGCAATAGTTGAACACTTGCCTGATCTCAATATGGAACGAGTGTACGAGGACCACGAAATGTTAGTAGACAATTTAATGTTATGGACGAGAGAATCGAAAAACAAAATATTGTTTGCCGAAAGGCCAGATAAGATTTCGTTGTTCCAAGCCCCCGAAAAGTTTCTACTGAACGATAACGATCGGGGTCGTTCAAGTGAACAAGACGATCACTCAAGGCAAGTGATGCTAGAAGAATTTTTCGGACAAAACGGTGCCGGAGCCAGTACTGTCCCTTCTGTATCCGGACATCCGGTCCCGCCGATGGAGGGACCACTTTACCTGAAGAACGACTCCAAAAAAGGATGGAAAAAAATTTACTGCGTCCTCCGACCATCAGGGCTTTACTACTCGCCCAAAGACAAGGTGAAAACGTTGAAAGAACTCGTCTGCTTGGCCACTTTCGACACCAACGAAGTATACATAGGTCTGAACTGGAAGAAGAAATACAAGTCCCCAACAGACTACTGCTTCGCTATCAAACATCCGAGACTCCAGCAACCGAAAAGCGTCAAATTTATCAAGTTCTTGTGCGCTGAAGACCAGCAGACAATGGAAAGATGGATGACAGCTATGCGAATAGCTAAGCATGGAAGGCAATTGCTTGAAAATTACCGGTCCTTAGTTGAGGAGCTCACTCAAGAAGATTTAGATCATCTTGCTCACGCGCGTTCTTGTTCCATTACTTCAATACCGACAAAAACGAACAACGGTGTGACAAACATAAATAGTCCAGCTCAATCAACGTCAAGCAACATGAGTGTTGCCAACTCGGACATTAGCAGCGGGAGACATTCGCGTGCTTCTTCATCGAGCTCGAGTGGTTGTCTATCGGATGGAGGTACTGCTTCTGAAAGCGCTTTCGACTGTGAATTTCCTATGGGTACAATAAAACGCAAACCATCTATGAAACCCAACATTCCGCTAACGTGGATGACTCGACAATTGAAAGAGATGGTGGAGAAAGACGATATTACTTCAGAGGCTGACGGTATCGGTGATTCGGGGACACTCACCAGGAAGCCCAGGAAGAATCGCTGTGATGACTCTACACTGAAGCGCCATCATAATATTGATTCAACGGATACTAGTGTGTATGGCACAGCTACTACCGTGAGCATTACAAGTACAGAAAGTCCAGTCCGAAAGGAGCCTGTTTCTCCGACGTACGATCACTATGACACAATAACTCGCGACAGCTCCTTCAGAAACAGTACAGACAATAGTTCATCTCTGTACGGATACACCACAATATACGACAAAGTGCAGAAACAACAAGCTGAATGTACCGTTGAAGATTTGCCTTTGCCACCGCCCCCTGTCGTTGCAGATGTCCCAGACGCAATGTTCAGTTCAACACTGAGCCTCGACAGCCTTCCTCCACCGCCGCCACCGCTGGACGCGATTGAGGATTTGTCGGGATCTCTACTAAGCCTTCCCCCACCACCACCCGAGCACGCCATGGAGGCCAACAGTGGTCGAGTGCAAGACATCGTCAGTCAACTGACGGCACAACAAATAGAACAAGCTTCGCGAGCTGGTCAATGCAGAGTTACAGCTCGAACAAGTGAGCCGACCCGTTCCTTCCCGAGGCAACCCTCTCTCGACAGCGTCCACTCGGAGGCTTCGCGTAGCTCGTCACTCCCATCTGACAAAAGCATTTATGGCCCGCAAAACGTCGCGTATGGGGCCTGCCTAGTCGAGTTGCAGACTAAAAAATTAAGCAGCCCCTCCATTCAAAAAAAGGTGATCACACAAACTGAACCAACGAAGGAGCGAACGGGTTCTATAAAAAAAGTTAATTTTGCCGATGACTTACCGACGTGCTCCGAAAAGAAGGTCAAAAAGATCTCGTTCAATCTCAAAGAGCAACCTCCGCTTTCGCCGCGGAAACCGCCCCCGCCGAAACGTAACGAAAGCACTCGACTTACATCACCGAAAAAGCTAGTCGATTCAAACAGTAATCCTCCAAAGGAATTTCTAAAAGATCTGCAGAGGGTGATGCGGAAAAAATGGCAAGTCGCACAAAAGTGTAAACTCGAACCGGCAACAACTCCGCACGAGGTACTTGGTTTCCGTGAATATCCACTTGCGGATGATTACAAAGAGACGAGCGTGTCGATGTGGGTGCAGGAGCACTACGGCAGCGGCGTGGAGGACCCCTTCTACGAGAACGTGTATAGTGCGGAACAGACGGCAGTGCGACGCGAGGAACCCAAGCCCATAAAGAAGCGTCCACCACCAGCACCGCCGCGACGGAGCGACTCCACGCACCTGTCCACGCACCCGCACGCCTCAGCCGTACAGCCCACCGCTTGA
Protein
MALVEEVGAGSTADDPEALLNEWLGELTVLTAGLNSTGDGSLALRPLEIVAPRIDTYRFSMANLEETQDADLDAILGELCALDSEYDEELSRVSSGFASGSKDRVPRTTEPCTVRQEKDNSDAASGITRTESPDNDSAFSDTVSMLSSESSASSSASAKCKALKLNLHQNQKDASFQQKADKIKLALERMREANIKKLFIKAFSMDGSSKSLLVDEKMTCGYVTRLLADKNHVNMEPKWAIVEHLPDLNMERVYEDHEMLVDNLMLWTRESKNKILFAERPDKISLFQAPEKFLLNDNDRGRSSEQDDHSRQVMLEEFFGQNGAGASTVPSVSGHPVPPMEGPLYLKNDSKKGWKKIYCVLRPSGLYYSPKDKVKTLKELVCLATFDTNEVYIGLNWKKKYKSPTDYCFAIKHPRLQQPKSVKFIKFLCAEDQQTMERWMTAMRIAKHGRQLLENYRSLVEELTQEDLDHLAHARSCSITSIPTKTNNGVTNINSPAQSTSSNMSVANSDISSGRHSRASSSSSSGCLSDGGTASESAFDCEFPMGTIKRKPSMKPNIPLTWMTRQLKEMVEKDDITSEADGIGDSGTLTRKPRKNRCDDSTLKRHHNIDSTDTSVYGTATTVSITSTESPVRKEPVSPTYDHYDTITRDSSFRNSTDNSSSLYGYTTIYDKVQKQQAECTVEDLPLPPPPVVADVPDAMFSSTLSLDSLPPPPPPLDAIEDLSGSLLSLPPPPPEHAMEANSGRVQDIVSQLTAQQIEQASRAGQCRVTARTSEPTRSFPRQPSLDSVHSEASRSSSLPSDKSIYGPQNVAYGACLVELQTKKLSSPSIQKKVITQTEPTKERTGSIKKVNFADDLPTCSEKKVKKISFNLKEQPPLSPRKPPPPKRNESTRLTSPKKLVDSNSNPPKEFLKDLQRVMRKKWQVAQKCKLEPATTPHEVLGFREYPLADDYKETSVSMWVQEHYGSGVEDPFYENVYSAEQTAVRREEPKPIKKRPPPAPPRRSDSTHLSTHPHASAVQPTA
Summary
Uniprot
EMBL
Proteomes
PRIDE
Interpro
SUPFAM
SSF54236
SSF54236
Gene 3D
ProteinModelPortal
PDB
4GMV
E-value=3.25404e-66,
Score=643
Ontologies
GO
PANTHER
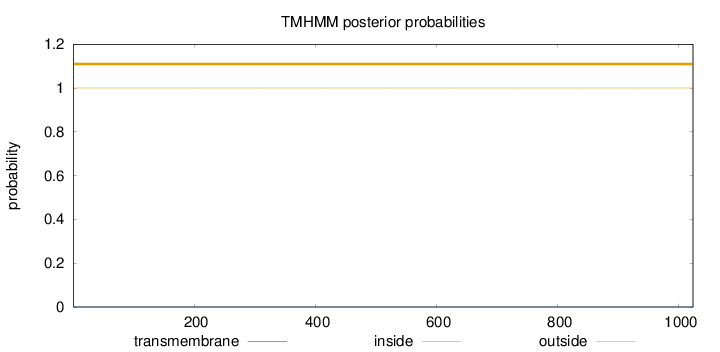
Topology
Length:
1023
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0.00023
Total prob of N-in:
0.00002
outside
1 - 1023
Population Genetic Test Statistics
Pi
1.664884
Theta
23.591613
Tajima's D
-2.364151
CLR
0.563895
CSRT
0.00179991000449977
Interpretation
Possibly Positive selection
