Gene
KWMTBOMO00332 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000529
Annotation
PREDICTED:_neutral_endopeptidase_24.11_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.845 Mitochondrial Reliability : 1.29
Sequence
CDS
ATGCGATACAGACAACAGTTTCGTAAGACTCTGACCAAGGTGGACTGGATGGATGATATGACTCGCCAAGAGGCTCTGGAGAAAGCTGATGCTATGGCTTCACACATCGCCTACCCCAGTGAAATGCTTGACAACAACAGACTCACCGAATTCTACTCTGGTCTGGAAATGTCATCGGAACACTTGATGGAGTCCGTGCTCAACTTGACATTGTTCACTACTGAGTACTTATTCGGTAAACTGAGGGAGCCCGTCAACAAGACTGACTGGGTCACCCACGGTCGCCCAGCTATCGTCAACGCTTTCTATTCGTCTATTGAGAACAGCATACAATTTCCGGCTGGTATCCTACAAGGAGCATTCTTCTCTGCCAAACGTCCTGCTTACATGAATTACGGTGCTATTGGTTTCGTCATTGGACATGAAATCACGCACGGTTTTGATGACCAGGGCCGGCAATTCGATAAAAACGGAAACCTTGTGGACTGGTGGCAGGAAATGACCAAAGAGAAGTATCTGGATAAAGCCAAATGTATCATCGATCAATACTCAAACTACACTGTTAAAGAAGTCGGATTGAAGTTGAATGGAGTGAACACACAAGGCGAGAACATTGCGGATAACGGAGGCATCAAAGAAGCTTACTACGCGTATCAAGCCTGGACTCACAGGCATGGTGAAGAAGCACGCCTGCCTGGATTGGAAAAATACAGTCCCCGACAGCTTTTCTGGCTTAGTGCTGCCAACACTTGGTGCGCAGTGTACCGCAATGAGGCAATCAAGTTGAGAATCACAACTGGATTCCATGCCCCCGGAAGATTCCGCGTCATTGGCCCCATGTCTAATATGGAAGAATTTGCCTCAGACTTCAAATGCCCTATGGGATCACCTATGAACCCTGACAAGAAATGCAAAGTGTGGTAG
Protein
MRYRQQFRKTLTKVDWMDDMTRQEALEKADAMASHIAYPSEMLDNNRLTEFYSGLEMSSEHLMESVLNLTLFTTEYLFGKLREPVNKTDWVTHGRPAIVNAFYSSIENSIQFPAGILQGAFFSAKRPAYMNYGAIGFVIGHEITHGFDDQGRQFDKNGNLVDWWQEMTKEKYLDKAKCIIDQYSNYTVKEVGLKLNGVNTQGENIADNGGIKEAYYAYQAWTHRHGEEARLPGLEKYSPRQLFWLSAANTWCAVYRNEAIKLRITTGFHAPGRFRVIGPMSNMEEFASDFKCPMGSPMNPDKKCKVW
Summary
Uniprot
Q9BLH1
H9ITE9
Q86RS4
A0A2A4JX35
A0A2U8JG00
A0A2A4JWQ5
+ More
A0A212F433 A0A1E1W8K7 A0A437B0E3 A0A2H1W5X7 S4NTA7 A0A194QVE0 A0A194PTK8 A0A1E1WV53 A0A154P8A9 A0A0L7R038 A0A310SLF2 A0A0C9S0X3 A0A0C9RKX4 A0A0M9A0V4 A0A088AKD7 E2C244 A0A2A3EL10 K7J3Q5 A0A151WK54 A0A2S2R687 A0A151J2K9 A0A195FJU4 J9JK87 A0A1I9WLF9 A0A151I329 A0A2H8TNG1 A0A1B6EX18 A0A158NUJ2 E9IRU9 A0A195C7U5 A0A026W1C7 F4WRD2 A0A2J7RR27 A0A3L8DWD5 A0A139WHP0 E2AFA5 A0A1W4W608 A0A1W4W5Z7 A0A172QPK9 A0A1B6CAF7 B0WKU3 A0A1B6DD22 A0A0J7NSM8 E0W1I2 A0A2J7RR19 A0A1Q3G120 A0A1Y1K718 A0A1Q3G115 A0A1Q3G159 A0A3Q0J997 A0A1Y1K673 A0A067RF34 A0A336MVN8 A0A182RNY0 A0A023EVW7 W5JAQ0 A0A182G3C1 A0A232F7R6 A0A182P9R2 A0A182J9J9 A0A182F8P8 U4U6M7 A0A182LWV4 A0A340TB26 A0A182T699 A0A182U1K2 N6UII9 A0A2M4CV03 A0A084VWM9 A0A182VC38 A0A182VRK4 A0A182Q5Q1 A0A182XV77 A0A182NFL8 A0A182JR91 A0A1S4FNM5 A0A146LYT5 A0A0K8SBS3 A0A1S4GDN6 A0A182HGU4 Q16UJ2 Q7PY82 K7J3Q4 A0A182X2S6 A0A1L8E2J5 A0A0K8TLN7 A0A182KMD2 A0A1L8E2C7 A0A2R7W332 A0A0A1XMX9 A0A1L8E257 J9JQQ7 A0A0L0CLG4 A0A1I8M295
A0A212F433 A0A1E1W8K7 A0A437B0E3 A0A2H1W5X7 S4NTA7 A0A194QVE0 A0A194PTK8 A0A1E1WV53 A0A154P8A9 A0A0L7R038 A0A310SLF2 A0A0C9S0X3 A0A0C9RKX4 A0A0M9A0V4 A0A088AKD7 E2C244 A0A2A3EL10 K7J3Q5 A0A151WK54 A0A2S2R687 A0A151J2K9 A0A195FJU4 J9JK87 A0A1I9WLF9 A0A151I329 A0A2H8TNG1 A0A1B6EX18 A0A158NUJ2 E9IRU9 A0A195C7U5 A0A026W1C7 F4WRD2 A0A2J7RR27 A0A3L8DWD5 A0A139WHP0 E2AFA5 A0A1W4W608 A0A1W4W5Z7 A0A172QPK9 A0A1B6CAF7 B0WKU3 A0A1B6DD22 A0A0J7NSM8 E0W1I2 A0A2J7RR19 A0A1Q3G120 A0A1Y1K718 A0A1Q3G115 A0A1Q3G159 A0A3Q0J997 A0A1Y1K673 A0A067RF34 A0A336MVN8 A0A182RNY0 A0A023EVW7 W5JAQ0 A0A182G3C1 A0A232F7R6 A0A182P9R2 A0A182J9J9 A0A182F8P8 U4U6M7 A0A182LWV4 A0A340TB26 A0A182T699 A0A182U1K2 N6UII9 A0A2M4CV03 A0A084VWM9 A0A182VC38 A0A182VRK4 A0A182Q5Q1 A0A182XV77 A0A182NFL8 A0A182JR91 A0A1S4FNM5 A0A146LYT5 A0A0K8SBS3 A0A1S4GDN6 A0A182HGU4 Q16UJ2 Q7PY82 K7J3Q4 A0A182X2S6 A0A1L8E2J5 A0A0K8TLN7 A0A182KMD2 A0A1L8E2C7 A0A2R7W332 A0A0A1XMX9 A0A1L8E257 J9JQQ7 A0A0L0CLG4 A0A1I8M295
Pubmed
11583934
19121390
22118469
23622113
26354079
20798317
+ More
20075255 27538518 21347285 21282665 24508170 21719571 30249741 18362917 19820115 20566863 28004739 24845553 24945155 20920257 23761445 26483478 28648823 23537049 24438588 25244985 17510324 26823975 12364791 26369729 20966253 25830018 26108605 25315136
20075255 27538518 21347285 21282665 24508170 21719571 30249741 18362917 19820115 20566863 28004739 24845553 24945155 20920257 23761445 26483478 28648823 23537049 24438588 25244985 17510324 26823975 12364791 26369729 20966253 25830018 26108605 25315136
EMBL
AB048208
BAB33300.1
BABH01007095
BABH01007096
AF413063
AAO21504.1
+ More
NWSH01000486 PCG76063.1 MH298329 AWK67618.1 PCG76064.1 AGBW02010446 OWR48508.1 GDQN01007734 JAT83320.1 RSAL01000227 RVE43942.1 ODYU01006558 SOQ48505.1 GAIX01012266 JAA80294.1 KQ461108 KPJ09289.1 KQ459594 KPI96089.1 GDQN01000159 JAT90895.1 KQ434824 KZC07350.1 KQ414672 KOC64187.1 KQ762520 OAD55759.1 GBYB01014266 JAG84033.1 GBYB01007566 JAG77333.1 KQ435774 KOX75065.1 GL452067 EFN77995.1 KZ288217 PBC32410.1 KQ983014 KYQ48238.1 GGMS01016230 MBY85433.1 KQ980368 KYN16324.1 KQ981522 KYN40631.1 ABLF02019556 KU932343 APA33979.1 KQ976533 KYM81493.1 GFXV01003870 MBW15675.1 GECZ01027204 JAS42565.1 ADTU01026577 ADTU01026578 ADTU01026579 ADTU01026580 ADTU01026581 GL765283 EFZ16711.1 KQ978143 KYM96720.1 KK107496 EZA49818.1 GL888285 EGI63203.1 NEVH01000611 PNF43277.1 QOIP01000004 RLU24088.1 KQ971343 KYB27446.1 GL439090 EFN67881.1 KU245762 AND99761.1 GEDC01026857 JAS10441.1 DS231976 EDS30066.1 GEDC01013803 JAS23495.1 LBMM01001992 KMQ95435.1 DS235870 EEB19488.1 PNF43276.1 GFDL01001550 JAV33495.1 GEZM01094022 JAV56000.1 GFDL01001557 JAV33488.1 GFDL01001511 JAV33534.1 GEZM01094021 JAV56001.1 KK852675 KDR18716.1 UFQS01003165 UFQT01003165 SSX15299.1 SSX34674.1 GAPW01000331 JAC13267.1 ADMH02001926 ETN60478.1 JXUM01040714 JXUM01040715 JXUM01040716 JXUM01040717 JXUM01040718 JXUM01040719 JXUM01040720 NNAY01000760 OXU26642.1 KB632006 ERL87973.1 AXCM01000914 APGK01034174 APGK01034175 APGK01034176 APGK01034177 APGK01034178 KB740904 ENN78462.1 GGFL01004490 MBW68668.1 ATLV01017670 KE525181 KFB42373.1 AXCN02000208 GDHC01006833 JAQ11796.1 GBRD01015302 JAG50524.1 AAAB01008987 APCN01005946 CH477621 EAT38188.1 EAA01126.5 GFDF01001146 JAV12938.1 GDAI01002525 JAI15078.1 GFDF01001260 JAV12824.1 KK854278 PTY14137.1 GBXI01001946 GBXI01001592 JAD12346.1 JAD12700.1 GFDF01001261 JAV12823.1 ABLF02033036 ABLF02033037 JRES01000236 KNC33072.1
NWSH01000486 PCG76063.1 MH298329 AWK67618.1 PCG76064.1 AGBW02010446 OWR48508.1 GDQN01007734 JAT83320.1 RSAL01000227 RVE43942.1 ODYU01006558 SOQ48505.1 GAIX01012266 JAA80294.1 KQ461108 KPJ09289.1 KQ459594 KPI96089.1 GDQN01000159 JAT90895.1 KQ434824 KZC07350.1 KQ414672 KOC64187.1 KQ762520 OAD55759.1 GBYB01014266 JAG84033.1 GBYB01007566 JAG77333.1 KQ435774 KOX75065.1 GL452067 EFN77995.1 KZ288217 PBC32410.1 KQ983014 KYQ48238.1 GGMS01016230 MBY85433.1 KQ980368 KYN16324.1 KQ981522 KYN40631.1 ABLF02019556 KU932343 APA33979.1 KQ976533 KYM81493.1 GFXV01003870 MBW15675.1 GECZ01027204 JAS42565.1 ADTU01026577 ADTU01026578 ADTU01026579 ADTU01026580 ADTU01026581 GL765283 EFZ16711.1 KQ978143 KYM96720.1 KK107496 EZA49818.1 GL888285 EGI63203.1 NEVH01000611 PNF43277.1 QOIP01000004 RLU24088.1 KQ971343 KYB27446.1 GL439090 EFN67881.1 KU245762 AND99761.1 GEDC01026857 JAS10441.1 DS231976 EDS30066.1 GEDC01013803 JAS23495.1 LBMM01001992 KMQ95435.1 DS235870 EEB19488.1 PNF43276.1 GFDL01001550 JAV33495.1 GEZM01094022 JAV56000.1 GFDL01001557 JAV33488.1 GFDL01001511 JAV33534.1 GEZM01094021 JAV56001.1 KK852675 KDR18716.1 UFQS01003165 UFQT01003165 SSX15299.1 SSX34674.1 GAPW01000331 JAC13267.1 ADMH02001926 ETN60478.1 JXUM01040714 JXUM01040715 JXUM01040716 JXUM01040717 JXUM01040718 JXUM01040719 JXUM01040720 NNAY01000760 OXU26642.1 KB632006 ERL87973.1 AXCM01000914 APGK01034174 APGK01034175 APGK01034176 APGK01034177 APGK01034178 KB740904 ENN78462.1 GGFL01004490 MBW68668.1 ATLV01017670 KE525181 KFB42373.1 AXCN02000208 GDHC01006833 JAQ11796.1 GBRD01015302 JAG50524.1 AAAB01008987 APCN01005946 CH477621 EAT38188.1 EAA01126.5 GFDF01001146 JAV12938.1 GDAI01002525 JAI15078.1 GFDF01001260 JAV12824.1 KK854278 PTY14137.1 GBXI01001946 GBXI01001592 JAD12346.1 JAD12700.1 GFDF01001261 JAV12823.1 ABLF02033036 ABLF02033037 JRES01000236 KNC33072.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
+ More
UP000076502 UP000053825 UP000053105 UP000005203 UP000008237 UP000242457 UP000002358 UP000075809 UP000078492 UP000078541 UP000007819 UP000078540 UP000005205 UP000078542 UP000053097 UP000007755 UP000235965 UP000279307 UP000007266 UP000000311 UP000192223 UP000002320 UP000036403 UP000009046 UP000079169 UP000027135 UP000075900 UP000000673 UP000069940 UP000215335 UP000075885 UP000075880 UP000069272 UP000030742 UP000075883 UP000075903 UP000075901 UP000075902 UP000019118 UP000030765 UP000075920 UP000075886 UP000076408 UP000075884 UP000075881 UP000075840 UP000008820 UP000007062 UP000076407 UP000075882 UP000037069 UP000095301
UP000076502 UP000053825 UP000053105 UP000005203 UP000008237 UP000242457 UP000002358 UP000075809 UP000078492 UP000078541 UP000007819 UP000078540 UP000005205 UP000078542 UP000053097 UP000007755 UP000235965 UP000279307 UP000007266 UP000000311 UP000192223 UP000002320 UP000036403 UP000009046 UP000079169 UP000027135 UP000075900 UP000000673 UP000069940 UP000215335 UP000075885 UP000075880 UP000069272 UP000030742 UP000075883 UP000075903 UP000075901 UP000075902 UP000019118 UP000030765 UP000075920 UP000075886 UP000076408 UP000075884 UP000075881 UP000075840 UP000008820 UP000007062 UP000076407 UP000075882 UP000037069 UP000095301
Interpro
IPR024079
MetalloPept_cat_dom_sf
+ More
IPR008753 Peptidase_M13_N
IPR042089 Peptidase_M13_dom_2
IPR018497 Peptidase_M13_C
IPR000718 Peptidase_M13
IPR042116 TypA/BipA_C
IPR011989 ARM-like
IPR036322 WD40_repeat_dom_sf
IPR038122 PFU_sf
IPR013535 PUL_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR015155 PFU
IPR016024 ARM-type_fold
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR008753 Peptidase_M13_N
IPR042089 Peptidase_M13_dom_2
IPR018497 Peptidase_M13_C
IPR000718 Peptidase_M13
IPR042116 TypA/BipA_C
IPR011989 ARM-like
IPR036322 WD40_repeat_dom_sf
IPR038122 PFU_sf
IPR013535 PUL_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR015155 PFU
IPR016024 ARM-type_fold
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
CDD
ProteinModelPortal
Q9BLH1
H9ITE9
Q86RS4
A0A2A4JX35
A0A2U8JG00
A0A2A4JWQ5
+ More
A0A212F433 A0A1E1W8K7 A0A437B0E3 A0A2H1W5X7 S4NTA7 A0A194QVE0 A0A194PTK8 A0A1E1WV53 A0A154P8A9 A0A0L7R038 A0A310SLF2 A0A0C9S0X3 A0A0C9RKX4 A0A0M9A0V4 A0A088AKD7 E2C244 A0A2A3EL10 K7J3Q5 A0A151WK54 A0A2S2R687 A0A151J2K9 A0A195FJU4 J9JK87 A0A1I9WLF9 A0A151I329 A0A2H8TNG1 A0A1B6EX18 A0A158NUJ2 E9IRU9 A0A195C7U5 A0A026W1C7 F4WRD2 A0A2J7RR27 A0A3L8DWD5 A0A139WHP0 E2AFA5 A0A1W4W608 A0A1W4W5Z7 A0A172QPK9 A0A1B6CAF7 B0WKU3 A0A1B6DD22 A0A0J7NSM8 E0W1I2 A0A2J7RR19 A0A1Q3G120 A0A1Y1K718 A0A1Q3G115 A0A1Q3G159 A0A3Q0J997 A0A1Y1K673 A0A067RF34 A0A336MVN8 A0A182RNY0 A0A023EVW7 W5JAQ0 A0A182G3C1 A0A232F7R6 A0A182P9R2 A0A182J9J9 A0A182F8P8 U4U6M7 A0A182LWV4 A0A340TB26 A0A182T699 A0A182U1K2 N6UII9 A0A2M4CV03 A0A084VWM9 A0A182VC38 A0A182VRK4 A0A182Q5Q1 A0A182XV77 A0A182NFL8 A0A182JR91 A0A1S4FNM5 A0A146LYT5 A0A0K8SBS3 A0A1S4GDN6 A0A182HGU4 Q16UJ2 Q7PY82 K7J3Q4 A0A182X2S6 A0A1L8E2J5 A0A0K8TLN7 A0A182KMD2 A0A1L8E2C7 A0A2R7W332 A0A0A1XMX9 A0A1L8E257 J9JQQ7 A0A0L0CLG4 A0A1I8M295
A0A212F433 A0A1E1W8K7 A0A437B0E3 A0A2H1W5X7 S4NTA7 A0A194QVE0 A0A194PTK8 A0A1E1WV53 A0A154P8A9 A0A0L7R038 A0A310SLF2 A0A0C9S0X3 A0A0C9RKX4 A0A0M9A0V4 A0A088AKD7 E2C244 A0A2A3EL10 K7J3Q5 A0A151WK54 A0A2S2R687 A0A151J2K9 A0A195FJU4 J9JK87 A0A1I9WLF9 A0A151I329 A0A2H8TNG1 A0A1B6EX18 A0A158NUJ2 E9IRU9 A0A195C7U5 A0A026W1C7 F4WRD2 A0A2J7RR27 A0A3L8DWD5 A0A139WHP0 E2AFA5 A0A1W4W608 A0A1W4W5Z7 A0A172QPK9 A0A1B6CAF7 B0WKU3 A0A1B6DD22 A0A0J7NSM8 E0W1I2 A0A2J7RR19 A0A1Q3G120 A0A1Y1K718 A0A1Q3G115 A0A1Q3G159 A0A3Q0J997 A0A1Y1K673 A0A067RF34 A0A336MVN8 A0A182RNY0 A0A023EVW7 W5JAQ0 A0A182G3C1 A0A232F7R6 A0A182P9R2 A0A182J9J9 A0A182F8P8 U4U6M7 A0A182LWV4 A0A340TB26 A0A182T699 A0A182U1K2 N6UII9 A0A2M4CV03 A0A084VWM9 A0A182VC38 A0A182VRK4 A0A182Q5Q1 A0A182XV77 A0A182NFL8 A0A182JR91 A0A1S4FNM5 A0A146LYT5 A0A0K8SBS3 A0A1S4GDN6 A0A182HGU4 Q16UJ2 Q7PY82 K7J3Q4 A0A182X2S6 A0A1L8E2J5 A0A0K8TLN7 A0A182KMD2 A0A1L8E2C7 A0A2R7W332 A0A0A1XMX9 A0A1L8E257 J9JQQ7 A0A0L0CLG4 A0A1I8M295
PDB
5V48
E-value=1.06139e-78,
Score=745
Ontologies
GO
PANTHER
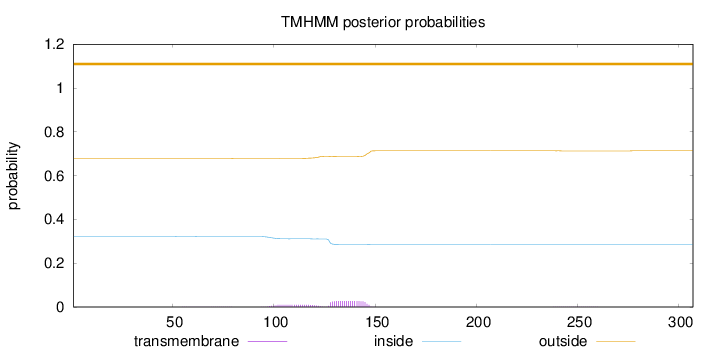
Topology
Length:
307
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.74965
Exp number, first 60 AAs:
0.00137
Total prob of N-in:
0.32173
outside
1 - 307
Population Genetic Test Statistics
Pi
4.904208
Theta
12.462965
Tajima's D
-1.333845
CLR
0
CSRT
0.0821458927053647
Interpretation
Uncertain
