Gene
KWMTBOMO00329
Pre Gene Modal
BGIBMGA000697
Annotation
PREDICTED:_probable_G-protein_coupled_receptor_52_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.975
Sequence
CDS
ATGCCAGCGAAGATGATGAATGCTAGAGGCGGTGCAAACGGGGCGGGTGAGGTGAGTGTGGCCGACAGTCCCCTGGCCACCTTTGAGTCACTCACGCAGGCTGCCGTTATAGCAGTGATGGGAGTTGCCATTGTTGTGTCCAATCTGCTAATAATCGCTGCATTCTTGAACTTCAAAGGTCTCTCAAACGAAGTGATAAACTACTATCTACTATCATTGGCTGTTGCGGATTTACTATGCGGCTTGCTTGTGGTACCACTATCAGTATACCCTGCGATAACAGGCCGCTGGATGTTCGGGGATCTCATGTGTAGACTTGCCGGCTACGTAGAAGTCACCTTGTGGTCCGTCTCAGTCTATACTTTCATGTGGATCTCCGTCGATAGATATCTTGCCGTCAGAAAACCTCTCCGATATGAGACGGTTAGTGCAACGGTGCAAACACGTACTCGCAGTCAATGCTGGATGGTATTCACTTGGATCTCGGCAGCCATGCTGTGTTGTCCGCCTTTACTTGGCTATAAAAAAGATGCAAACTTCGACAAGGAAACTTTCATATGCATGCTTGATTGGGGCACTACATACGCCTACACTGCCACTCTAGGTATCCTTGTGCTGGGACCTAGCGTTATTTCAATTGTTTACAACTACTTTTACATTTTTTCGATGAAACGGAAACTGCATAGCGGAGTGCCTATTCATGATAAGGAATACGCGACTGCTTTGGCTGAGAACCTCGCTAACCCTAGCCACTGGATGAGTTTCGTTTTGGTGTCCGTGTTCTGGTTGAGTTGGGCTCCGTATGCCGGTGTGAGAATGTACGAATACGTCACCAATCAAGAAATCAAAATTCCAATGTTACATTTCGGCATGGTTTGGCTTGGTATTATGAATTCGTTTTGGAAAATTATCATTTTGATTTCACTCAGCCCCCAGTTCCGGCTTGCGCTTCGAATTTTGTGTTTGACTGCATGCTGCCGCACCAAAGGACGCCTGCAAGCCGAACTCATCGGTATGGATAATGATGACTGA
Protein
MPAKMMNARGGANGAGEVSVADSPLATFESLTQAAVIAVMGVAIVVSNLLIIAAFLNFKGLSNEVINYYLLSLAVADLLCGLLVVPLSVYPAITGRWMFGDLMCRLAGYVEVTLWSVSVYTFMWISVDRYLAVRKPLRYETVSATVQTRTRSQCWMVFTWISAAMLCCPPLLGYKKDANFDKETFICMLDWGTTYAYTATLGILVLGPSVISIVYNYFYIFSMKRKLHSGVPIHDKEYATALAENLANPSHWMSFVLVSVFWLSWAPYAGVRMYEYVTNQEIKIPMLHFGMVWLGIMNSFWKIIILISLSPQFRLALRILCLTACCRTKGRLQAELIGMDNDD
Summary
Uniprot
H9ITW7
A0A212FHG5
R9UUE9
A0A2A4JVK3
A0A386QW71
A0A2H1VKY5
+ More
A0A0N0PB68 A0A0L7K4A0 A0A194Q706 A0A1B0GL43 A0A182LTR6 A0A182FDA8 A0A182NGY9 A0A182LIZ8 A0A182HWP9 D6WWZ1 A0A182Y1C6 A0A182PHQ1 A0A182UWB3 A0A182WIF6 A0A182XMU3 Q7PNR0 A0A182RL87 B4N480 B3M921 W5J530 A0A182Q8A5 A0A3B0JW73 A0A0A1XPQ6 W8B3I2 Q29ED2 A0A0A1XMC0 I2FKF3 B4HU56 B3NC31 B4QQK9 A0A1W4VAX9 Q9VZD1 B4PHU9 A0A1B0FNB8 A0A1A9ZS25 Q8T0P6 B4LGW7 A0A1A9YEH5 A0A1B0B7W3 A0A0M5IZ97 A0A182JDQ9 B4KWT0 A0A1A9WG10 A0A2J7RP62 B4IZP3 Q8MR64 A0A1I8MU74 N6TTM1 A0A1I8P5T9 B4K3N9 A0A2P8YJY8 Q17PM8 B0WIL2 A0A1B6E426 A0A1W4XSR8 A0A084W4A5 A0A1B6MP05 A0A195D3B3 F4WDX3 A0A195BAZ9 A0A158P2F6 A0A195FIV5 A0A3L8DYW2 E9IIV3 A0A088AS18 A0A2A3E8B1 V9IBB6 A0A154PFG8 A0A151WTY5 A0A0L7QTV1 A0A0N0BF00 E2AZZ1 E0W215 A0A336LKQ4 A0A151J9J1 A0A0T6BC88 E2CA46 A0A182GBT7 A0A224XG97 T1IE08 B4H1V5 A0A1S3D9Q8 A0A1B0D5T3 A0A1B6J175 A0A1J1IBP9 A0A034V829 A0A0A1WQR5 A0A1A9V486 A0A0J7KNM0 K7J7S3 A0A2S2QC33 A0A0N8A808
A0A0N0PB68 A0A0L7K4A0 A0A194Q706 A0A1B0GL43 A0A182LTR6 A0A182FDA8 A0A182NGY9 A0A182LIZ8 A0A182HWP9 D6WWZ1 A0A182Y1C6 A0A182PHQ1 A0A182UWB3 A0A182WIF6 A0A182XMU3 Q7PNR0 A0A182RL87 B4N480 B3M921 W5J530 A0A182Q8A5 A0A3B0JW73 A0A0A1XPQ6 W8B3I2 Q29ED2 A0A0A1XMC0 I2FKF3 B4HU56 B3NC31 B4QQK9 A0A1W4VAX9 Q9VZD1 B4PHU9 A0A1B0FNB8 A0A1A9ZS25 Q8T0P6 B4LGW7 A0A1A9YEH5 A0A1B0B7W3 A0A0M5IZ97 A0A182JDQ9 B4KWT0 A0A1A9WG10 A0A2J7RP62 B4IZP3 Q8MR64 A0A1I8MU74 N6TTM1 A0A1I8P5T9 B4K3N9 A0A2P8YJY8 Q17PM8 B0WIL2 A0A1B6E426 A0A1W4XSR8 A0A084W4A5 A0A1B6MP05 A0A195D3B3 F4WDX3 A0A195BAZ9 A0A158P2F6 A0A195FIV5 A0A3L8DYW2 E9IIV3 A0A088AS18 A0A2A3E8B1 V9IBB6 A0A154PFG8 A0A151WTY5 A0A0L7QTV1 A0A0N0BF00 E2AZZ1 E0W215 A0A336LKQ4 A0A151J9J1 A0A0T6BC88 E2CA46 A0A182GBT7 A0A224XG97 T1IE08 B4H1V5 A0A1S3D9Q8 A0A1B0D5T3 A0A1B6J175 A0A1J1IBP9 A0A034V829 A0A0A1WQR5 A0A1A9V486 A0A0J7KNM0 K7J7S3 A0A2S2QC33 A0A0N8A808
Pubmed
19121390
22118469
24023771
26354079
26227816
20966253
+ More
18054377 18025266 18362917 18316733 20068045 19820115 21843505 23604020 25244985 12364791 17994087 18057021 20920257 23761445 25830018 24495485 15632085 22936249 10731132 12537568 12537572 12537573 12537574 15987944 16110336 17569856 17569867 26109357 26109356 17550304 25315136 23537049 29403074 17510324 24438588 21719571 21347285 30249741 21282665 20798317 20566863 26483478 25348373 20075255
18054377 18025266 18362917 18316733 20068045 19820115 21843505 23604020 25244985 12364791 17994087 18057021 20920257 23761445 25830018 24495485 15632085 22936249 10731132 12537568 12537572 12537573 12537574 15987944 16110336 17569856 17569867 26109357 26109356 17550304 25315136 23537049 29403074 17510324 24438588 21719571 21347285 30249741 21282665 20798317 20566863 26483478 25348373 20075255
EMBL
BABH01007086
AGBW02008503
OWR53178.1
KC715734
AGN74919.1
NWSH01000486
+ More
PCG76067.1 MG596302 AYE54242.1 ODYU01002947 SOQ41092.1 KQ461033 KPJ09995.1 JTDY01010782 KOB58020.1 KQ459581 KPI99175.1 AJWK01034178 AXCM01001152 APCN01001522 BK005875 KQ971361 DAA64512.1 EFA09248.1 AAAB01008960 EAA11751.2 CH964095 EDW78954.1 CH902618 EDV38965.2 KPU77774.1 KPU77775.1 KPU77776.1 ADMH02002072 ETN59587.1 AXCN02000350 AXCN02000351 OUUW01000002 SPP77616.1 GBXI01001734 JAD12558.1 GAMC01014948 GAMC01014947 JAB91608.1 CH379070 EAL30129.1 GBXI01001823 JAD12469.1 AB720743 BAM15638.1 CH480817 EDW50477.1 CH954178 EDV50919.1 CM000363 CM002912 EDX09215.1 KMY97589.1 AE014296 AJ786412 AAF47893.1 AAX52732.1 AAX52733.1 CAH10219.1 CM000159 EDW93408.1 KRK01168.1 KRK01169.1 KRK01170.1 KRK01171.1 CCAG010000497 AY069139 AAL39284.1 CH940647 EDW68437.1 JXJN01009731 CP012525 ALC45081.1 CH933809 EDW17527.1 NEVH01002140 PNF42614.1 CH916366 EDV95628.1 AY122100 AAM52612.1 APGK01019213 APGK01019214 KB740096 KB632053 ENN81408.1 ERL88340.1 CH924146 EDV90449.1 PYGN01000541 PSN44569.1 CH477190 EAT48724.1 DS231949 EDS28535.1 GEDC01004634 JAS32664.1 ATLV01020303 KE525297 KFB45049.1 GEBQ01002382 JAT37595.1 KQ976885 KYN07400.1 GL888096 EGI67605.1 KQ976530 KYM81708.1 ADTU01007305 KQ981523 KYN40197.1 QOIP01000002 RLU25660.1 GL763562 EFZ19487.1 KZ288329 PBC27930.1 JR037728 AEY57937.1 KQ434893 KZC10626.1 KQ982753 KYQ51237.1 KQ414736 KOC62092.1 KQ435816 KOX72634.1 GL444289 EFN60994.1 DS235873 EEB19609.1 UFQS01000004 UFQT01000004 SSW96852.1 SSX17239.1 KQ979404 KYN21687.1 LJIG01002006 KRT84934.1 GL453920 EFN75201.1 JXUM01053225 JXUM01053226 JXUM01053227 KQ561768 KXJ77598.1 GFTR01004921 JAW11505.1 ACPB03000244 CH479203 EDW30307.1 AJVK01000300 GECU01014846 JAS92860.1 CVRI01000043 CRK95865.1 GAKP01019481 JAC39471.1 GBXI01013301 JAD00991.1 LBMM01005059 KMQ91846.1 AAZX01011149 GGMS01006102 MBY75305.1 GDIP01173473 LRGB01000687 JAJ49929.1 KZS16875.1
PCG76067.1 MG596302 AYE54242.1 ODYU01002947 SOQ41092.1 KQ461033 KPJ09995.1 JTDY01010782 KOB58020.1 KQ459581 KPI99175.1 AJWK01034178 AXCM01001152 APCN01001522 BK005875 KQ971361 DAA64512.1 EFA09248.1 AAAB01008960 EAA11751.2 CH964095 EDW78954.1 CH902618 EDV38965.2 KPU77774.1 KPU77775.1 KPU77776.1 ADMH02002072 ETN59587.1 AXCN02000350 AXCN02000351 OUUW01000002 SPP77616.1 GBXI01001734 JAD12558.1 GAMC01014948 GAMC01014947 JAB91608.1 CH379070 EAL30129.1 GBXI01001823 JAD12469.1 AB720743 BAM15638.1 CH480817 EDW50477.1 CH954178 EDV50919.1 CM000363 CM002912 EDX09215.1 KMY97589.1 AE014296 AJ786412 AAF47893.1 AAX52732.1 AAX52733.1 CAH10219.1 CM000159 EDW93408.1 KRK01168.1 KRK01169.1 KRK01170.1 KRK01171.1 CCAG010000497 AY069139 AAL39284.1 CH940647 EDW68437.1 JXJN01009731 CP012525 ALC45081.1 CH933809 EDW17527.1 NEVH01002140 PNF42614.1 CH916366 EDV95628.1 AY122100 AAM52612.1 APGK01019213 APGK01019214 KB740096 KB632053 ENN81408.1 ERL88340.1 CH924146 EDV90449.1 PYGN01000541 PSN44569.1 CH477190 EAT48724.1 DS231949 EDS28535.1 GEDC01004634 JAS32664.1 ATLV01020303 KE525297 KFB45049.1 GEBQ01002382 JAT37595.1 KQ976885 KYN07400.1 GL888096 EGI67605.1 KQ976530 KYM81708.1 ADTU01007305 KQ981523 KYN40197.1 QOIP01000002 RLU25660.1 GL763562 EFZ19487.1 KZ288329 PBC27930.1 JR037728 AEY57937.1 KQ434893 KZC10626.1 KQ982753 KYQ51237.1 KQ414736 KOC62092.1 KQ435816 KOX72634.1 GL444289 EFN60994.1 DS235873 EEB19609.1 UFQS01000004 UFQT01000004 SSW96852.1 SSX17239.1 KQ979404 KYN21687.1 LJIG01002006 KRT84934.1 GL453920 EFN75201.1 JXUM01053225 JXUM01053226 JXUM01053227 KQ561768 KXJ77598.1 GFTR01004921 JAW11505.1 ACPB03000244 CH479203 EDW30307.1 AJVK01000300 GECU01014846 JAS92860.1 CVRI01000043 CRK95865.1 GAKP01019481 JAC39471.1 GBXI01013301 JAD00991.1 LBMM01005059 KMQ91846.1 AAZX01011149 GGMS01006102 MBY75305.1 GDIP01173473 LRGB01000687 JAJ49929.1 KZS16875.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000037510
UP000053268
+ More
UP000092461 UP000075883 UP000069272 UP000075884 UP000075882 UP000075840 UP000007266 UP000076408 UP000075885 UP000075903 UP000075920 UP000076407 UP000007062 UP000075900 UP000007798 UP000007801 UP000000673 UP000075886 UP000268350 UP000001819 UP000001292 UP000008711 UP000000304 UP000192221 UP000000803 UP000002282 UP000092444 UP000092445 UP000008792 UP000092443 UP000092460 UP000092553 UP000075880 UP000009192 UP000091820 UP000235965 UP000001070 UP000095301 UP000019118 UP000030742 UP000095300 UP000245037 UP000008820 UP000002320 UP000192223 UP000030765 UP000078542 UP000007755 UP000078540 UP000005205 UP000078541 UP000279307 UP000005203 UP000242457 UP000076502 UP000075809 UP000053825 UP000053105 UP000000311 UP000009046 UP000078492 UP000008237 UP000069940 UP000249989 UP000015103 UP000008744 UP000079169 UP000092462 UP000183832 UP000078200 UP000036403 UP000002358 UP000076858
UP000092461 UP000075883 UP000069272 UP000075884 UP000075882 UP000075840 UP000007266 UP000076408 UP000075885 UP000075903 UP000075920 UP000076407 UP000007062 UP000075900 UP000007798 UP000007801 UP000000673 UP000075886 UP000268350 UP000001819 UP000001292 UP000008711 UP000000304 UP000192221 UP000000803 UP000002282 UP000092444 UP000092445 UP000008792 UP000092443 UP000092460 UP000092553 UP000075880 UP000009192 UP000091820 UP000235965 UP000001070 UP000095301 UP000019118 UP000030742 UP000095300 UP000245037 UP000008820 UP000002320 UP000192223 UP000030765 UP000078542 UP000007755 UP000078540 UP000005205 UP000078541 UP000279307 UP000005203 UP000242457 UP000076502 UP000075809 UP000053825 UP000053105 UP000000311 UP000009046 UP000078492 UP000008237 UP000069940 UP000249989 UP000015103 UP000008744 UP000079169 UP000092462 UP000183832 UP000078200 UP000036403 UP000002358 UP000076858
Pfam
PF00001 7tm_1
ProteinModelPortal
H9ITW7
A0A212FHG5
R9UUE9
A0A2A4JVK3
A0A386QW71
A0A2H1VKY5
+ More
A0A0N0PB68 A0A0L7K4A0 A0A194Q706 A0A1B0GL43 A0A182LTR6 A0A182FDA8 A0A182NGY9 A0A182LIZ8 A0A182HWP9 D6WWZ1 A0A182Y1C6 A0A182PHQ1 A0A182UWB3 A0A182WIF6 A0A182XMU3 Q7PNR0 A0A182RL87 B4N480 B3M921 W5J530 A0A182Q8A5 A0A3B0JW73 A0A0A1XPQ6 W8B3I2 Q29ED2 A0A0A1XMC0 I2FKF3 B4HU56 B3NC31 B4QQK9 A0A1W4VAX9 Q9VZD1 B4PHU9 A0A1B0FNB8 A0A1A9ZS25 Q8T0P6 B4LGW7 A0A1A9YEH5 A0A1B0B7W3 A0A0M5IZ97 A0A182JDQ9 B4KWT0 A0A1A9WG10 A0A2J7RP62 B4IZP3 Q8MR64 A0A1I8MU74 N6TTM1 A0A1I8P5T9 B4K3N9 A0A2P8YJY8 Q17PM8 B0WIL2 A0A1B6E426 A0A1W4XSR8 A0A084W4A5 A0A1B6MP05 A0A195D3B3 F4WDX3 A0A195BAZ9 A0A158P2F6 A0A195FIV5 A0A3L8DYW2 E9IIV3 A0A088AS18 A0A2A3E8B1 V9IBB6 A0A154PFG8 A0A151WTY5 A0A0L7QTV1 A0A0N0BF00 E2AZZ1 E0W215 A0A336LKQ4 A0A151J9J1 A0A0T6BC88 E2CA46 A0A182GBT7 A0A224XG97 T1IE08 B4H1V5 A0A1S3D9Q8 A0A1B0D5T3 A0A1B6J175 A0A1J1IBP9 A0A034V829 A0A0A1WQR5 A0A1A9V486 A0A0J7KNM0 K7J7S3 A0A2S2QC33 A0A0N8A808
A0A0N0PB68 A0A0L7K4A0 A0A194Q706 A0A1B0GL43 A0A182LTR6 A0A182FDA8 A0A182NGY9 A0A182LIZ8 A0A182HWP9 D6WWZ1 A0A182Y1C6 A0A182PHQ1 A0A182UWB3 A0A182WIF6 A0A182XMU3 Q7PNR0 A0A182RL87 B4N480 B3M921 W5J530 A0A182Q8A5 A0A3B0JW73 A0A0A1XPQ6 W8B3I2 Q29ED2 A0A0A1XMC0 I2FKF3 B4HU56 B3NC31 B4QQK9 A0A1W4VAX9 Q9VZD1 B4PHU9 A0A1B0FNB8 A0A1A9ZS25 Q8T0P6 B4LGW7 A0A1A9YEH5 A0A1B0B7W3 A0A0M5IZ97 A0A182JDQ9 B4KWT0 A0A1A9WG10 A0A2J7RP62 B4IZP3 Q8MR64 A0A1I8MU74 N6TTM1 A0A1I8P5T9 B4K3N9 A0A2P8YJY8 Q17PM8 B0WIL2 A0A1B6E426 A0A1W4XSR8 A0A084W4A5 A0A1B6MP05 A0A195D3B3 F4WDX3 A0A195BAZ9 A0A158P2F6 A0A195FIV5 A0A3L8DYW2 E9IIV3 A0A088AS18 A0A2A3E8B1 V9IBB6 A0A154PFG8 A0A151WTY5 A0A0L7QTV1 A0A0N0BF00 E2AZZ1 E0W215 A0A336LKQ4 A0A151J9J1 A0A0T6BC88 E2CA46 A0A182GBT7 A0A224XG97 T1IE08 B4H1V5 A0A1S3D9Q8 A0A1B0D5T3 A0A1B6J175 A0A1J1IBP9 A0A034V829 A0A0A1WQR5 A0A1A9V486 A0A0J7KNM0 K7J7S3 A0A2S2QC33 A0A0N8A808
PDB
6IBL
E-value=9.10809e-19,
Score=229
Ontologies
KEGG
GO
GO:0016021
GO:0008502
GO:0004930
GO:0071482
GO:0007186
GO:0008020
GO:0005887
GO:0007602
GO:0071329
GO:0008306
GO:0046959
GO:0035075
GO:0009267
GO:0007189
GO:0035100
GO:0043410
GO:0048149
GO:0001588
GO:0008227
GO:0009744
GO:0045471
GO:0042594
GO:0007018
GO:0030286
GO:0006813
GO:0006810
GO:0009072
GO:0007169
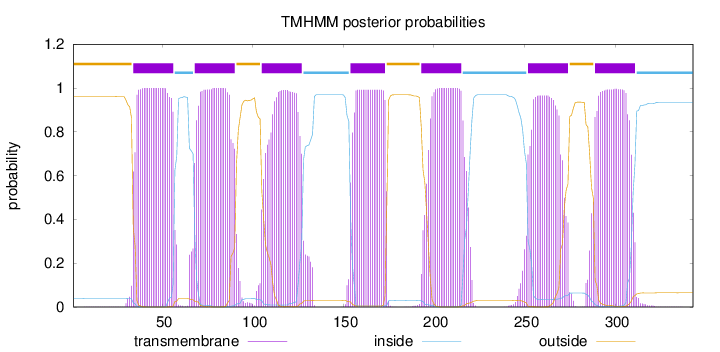
Topology
Length:
343
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
153.28096
Exp number, first 60 AAs:
22.91816
Total prob of N-in:
0.03927
POSSIBLE N-term signal
sequence
outside
1 - 33
TMhelix
34 - 56
inside
57 - 67
TMhelix
68 - 90
outside
91 - 104
TMhelix
105 - 127
inside
128 - 153
TMhelix
154 - 173
outside
174 - 192
TMhelix
193 - 215
inside
216 - 251
TMhelix
252 - 274
outside
275 - 288
TMhelix
289 - 311
inside
312 - 343
Population Genetic Test Statistics
Pi
19.251643
Theta
18.374516
Tajima's D
0.223481
CLR
0.189243
CSRT
0.428978551072446
Interpretation
Uncertain
