Gene
KWMTBOMO00319 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000691
Annotation
PREDICTED:_collagen_alpha-1(I)_chain-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.511
Sequence
CDS
ATGGCGCTCCTAAGCCAACGTCGAGCCGGTCTTGCTTCTCATTTTGCTATAATCATATGTTACTTTTGTGTATTGCAGTCTACAACTTCTCATGAAATTTCATCTGGTTATTATTTGATTTGCCGCGACACAAACACGCAGCTATGTGTTACTTGGCTAATCCAATACTCCCAAGCTATGAAGTTGGCTACAGAATTGAGTACAAAGAAAGTATCAACAGATGTTTGTGGATTCAGCCAATTGAAGCCTATGGGGCAAAATCGGTCATACGTAGAGGTATACCCGATTGACGAAAGTAAGCCGGTTATGCGCTGTCCTATAAAAAGTTTGGTGGTGAAGTACAACAAAGATCTGCTTAATTTGAAGAGCATACATGAAATACACAGAGTCCGTCGTCAAATTCCCTACGCATTACGAGGACGTTACAGGGGACAGACCCAATCTCAGTATCTGGCCATAGACCATGGAAACGGAAAAGATGATGGAAAAGCCGAAGCGCAATCAGCTGCTGACTCCTCTAGAGCTAGCGTCAGTGGAACGAGCGGAATGGGTCAAGCACAAAGTCAATCCATGTATGACCCGACCTGCGATGACTGTGTCGGCCGATCTACTGGCATCGATACCTCAAAACAAAAACCGTTGGGAGATTCTCACGGCGGCATTCCATCAGGATTACCTTCAGGAGCGATCGGCACACCAGGAAACAATTTACCCTTTGATCCAACAGGGGGAAGTCGATTAACAGGAAATGGAGGTTACGGGCCTAATGAAAATGGCGAACGTTATGGTCCTGGAAGTACTGGAATCAATGGTGCTTATAGACCACCGGGCGCTGGAGGAACTTATGGTCCTAATCAAATTGGGAACGGAGCTTTTAATCCAGCTCTAAATGGTATTCCAGGTATTCGTGTACAGGGACCAAATACAGGTTATGGAATAAGCCCTGGGGGTGGTTACACCGGATCATCTCCTGGGTTGGGTAATCAGTATTTGCCACCTTCAACTAGTACTAATTACGAACCAGTAAAAGGCAGCTATGGTCCAAGGCAAAATGTTCCTGATGGGTCTATAAATGGCTACCTTCCTGTACCTAGTGGTAGTAATGTATACGGGCCAAATCCTAACTCTGGCTCTGGTCACGGAATCGTTTCCAATATACCAGGAGGTGGCTATGGTCAAAATGGAAACTACGGACCCAACCAAAATGGAAATATGCCTTATGGTACAAACCAAGTCCCTTCACAAGGTACGAAAACGCCTGCTCATGGAGGAGGTCCAAGTGGGATCAATACGGGGTTAAGTTGGCCATCCACAGGAACATTTACACGTCCGGGATCTCAGAATACACCTAATGGACCAAGTTCTCAAGGCAACGCAAACGAACCTCATAATTTACTAGCCCCTAATGCTGTTTACGGACCTGGTGGCAATAGACCATATCAAACGGATGGCCAAAGTAACTACCAAACAAAACCAAATAACGCGTACGGAACTACTGGTGGATACACTCCTGGATCGAATGGTCCGTTTAATCCAGCTGACATAGGTACTAACAATAATGCAAACAAATATGGTCAGTTAAATCCAAATAATCAGAACGGCCTACAAAGTCCCGGTACTTACAGCCCTACGAGTACCGGACAAAACCACATGCCTGGAGTTATCGGTAGCCAAAATGGCCTACAAGGACCTGGAGGACAGTATGCGCCTACAAATACCGATGGGCAAATACCTGGAGTCCAATATAATCCTGGGAGCACAGTAGGTCATAATGGCTTGCCTATTCCTGGAGGTCTGTATAATTCTGGAATTGGAGGAGGTCCAAACAGTTTTCCTGGAACTCCAGGACAGTACCAACCTGGATCCACAGATGGTCATAATATTGTAACAGGCCCTGGAGTTCAATACAGACCAGATATCGTTAGTGGTCAAAACGGCTTACAAGGTCCCGGTAGTCAATATGGAGGTGGTCAAAGTAGGAATCAAGAGCCCGGAGGTCAATATGCTCCTGGAAATCAAAACATTATGCAAAGGCCAAGTGGACAGTATGGACCCGATAGTGCTGTTGGACAAGGTATTCAAGAACCAGGAAATATGTATGATTCTAGACCAAATAATCAAAGTCCTCAATATGGACCTGGAATCGGAATCGGTCCGAACGGATTACAAGGGCCAACTGGTCAGTATAGGCCTGGATCTTCCGGAGGTCAAAATGTTGTATCAGGCCCTGGAGGACAATATGGTAGTTCTGGAACTCAATATGGTCCAGGTGGTCAAAATGGAATTCAAGGTCCTAGTAGTCAATATGGTCCAGGTGGTCAAAACGGAATTCAAGTTCCTGGAACTCAATATGGTCCAGGGGGACAAAATGGAATACAAGTTCCCGGAACTGAGTATGGTCCAGGAGGTCAAAGAGGAATCCAAGGTCCTGGAACTCTGTATGGTCCTAATGGACAAAATGGAATCCAAAGTCCAGGAACTCAGTATGGTCCAGGGGGACAAATAGGAATTCAAGCACCTGGAACTCAATATGGACCAGGTGAGCAAAATGGAATCCAAAGTCCTGGAACTAAGACTGGTCCAGAAGGACAACTTGGAAACCAAGGGCCAGGGACACAGTATGGTCCAGGTGGACAATATGGAATCCAAGGGCCAGGAAATCAGTATGTCCCCGGCGGACAAGGTGGAATCCAAGCTCCTGTAAGTCAATATGATAATGTTCAAGGTGGAATTCGAGGACCAGGAACTCAGTATGGTCCAAGCGGACAAAGTGGCCTTCAAGGGCCCGGAGGCCAAAATGGCATACAATATCCAGGAGGAAGTTATGGCACCGGCGTCCAAAGTGGTTTGCAGAAACCAGGTGGCGTTTATGGTTCTGGGTTAAATGGAATCCAAAACAATGGCGTACCTGGAACGGGTGATAATTATGTTCCTGGAGGCCAAAGTAATGTGCCAGGTGGTTACGGCATCTCTAATAGACCTTTTAATCTTGATGGTACTGGGAGTCAACAAGGTCCATATAACCCATCATTACCAGGACAAAATGGACCATATGGGACTAATATGGTAGGTCAAGGATCAGGTTCAATTAGTCCTGGCACTACTGGCTTAGGGAACACAGGTAATCAAAATGGACAGTACGGACCTAGTGCTGGACCTTACCCTAACCAATATGGTAATGACTTATCCAACACGGGAGGTGGTTCTGGACAGTATGCCAATGTTCCTTATGGATACGGTAATAATCAAGCTGGTGCACCACAGAATGTTGTCGATCCAAATTCAGCTCTTTTGATAGATGGTGACGATTCTGCAGCTGAAGCTAGCGTTTCTCAAGCTTCAAACGGTACAACGGCTATAGCTTCCTCAAAAGGAGGAAATGATAAAGGACGTGCTCAAACACATGTGCAAGGAGCTTATACTGGCGGTGGCTCATTTTCTGCTCAAGCCGAAATTAGTGGATCCGAGACAGGTTCCATCTTGACTGGTTCGCAAAGCGAAGGAGTGATGCATTCTTCCAATACACAAGTACAGGGTAGCTTGAAAGGTGGCATGGCGGACGCTCAAGCTCGTGGTCCAGGTAGCACTTCGTCTCAAGCTCAGATTGGTTTTACACCGTATACACATGGAGATGCATCTCATGATGTACAAAAATCTCCATTTGTGGGCGGGGGAACTGCAGCTGCACAATCAAGTGGTCGAACGGGCTCCTCACAATCACAATTACGTGGAACATTTAAATATGGAATTACTTATAACGGAGGAGCTCAAGCAGGTGCCAGCTTAGACAAAGACACAGTCTTTTCTAATAGACTTCCATTTGATAAAATTGATGTTTATGATGAAAAAAACAAAAATATTAACATCGAGACGACGGAGAAATCATTAGACTTGTTACCTACTGAATCGACTGAAATCCCACTAACTACGGAACACTTGAGCTATGATATTCCTTCAACTACACCGTTGGATATCAGTGCAAACACGGAAAAAATAGATCCAGTATATTCTGAAAATAACGACGAATATGGGTCACATTCTCACTCTGATCATCACAAAACTGAATTTGCGGCATCTACACCAACTCGTTCACCTCCCGTTGAAAGTAGACGCTCTTTTCAGTCCGCTTATGGCACTAATAGTGGAGATTATGATTATACCACTGATAAAGAAGAAAACCAACCTGAGGAATATGATATGGATGACGGGTTTGCACCTGAAGGTACAGACACAAGTGATAACGATCAAGGAGAATTTGCAAATTATAATGAATTTTCTAGAGACTCAGAAACGCACCAATCTCTTCATTCGCCGAAGCGAAAAGGCCTGGAAGTAAGGCAAACAACAGGCGGCAACACGCAACATATTGTCTTGGGGGCACTTCGAGACCAAAATGCAGAGATTACACAGAGAAACTCTGAACGTCCTGATGAGAGTAAAATATATCAGCCTGGTGAGCGTGTACCAGGTACTGGCGGTTATACAATACCAGTTGGATTCACAGGCAGCGTTAAATCGGTTGCTTCCAAAGATAAAACTTACGTAGTAGGGTCAAAAGATTCACCATCTCAAGCGCAGACTGTATCCCTTACTCCTGGCTCTGGTAAAATAAAATATACTTATCCAACTACACACACCAGAAATGTGAGTCCTAAAGATTTGCGGTCACTTTACAATGTTAAACCAGATGATAATAATCGATACGTATCTGTTTCAAAATCAGTAACACGCGATTTAGACAGTGATAATAATATAAGGAAACAATATTCACACACCTACTATACAAAATCGTCATCTTGTGGATATTTTACTTTTACTTGTAATATGGTGAGTGGTGCTGAAGGCAAAAAAAAGGTTTGTAAGCCAAAAATTCCAACAAATCCTGATGGAACTCCTATGAGATGTTAA
Protein
MALLSQRRAGLASHFAIIICYFCVLQSTTSHEISSGYYLICRDTNTQLCVTWLIQYSQAMKLATELSTKKVSTDVCGFSQLKPMGQNRSYVEVYPIDESKPVMRCPIKSLVVKYNKDLLNLKSIHEIHRVRRQIPYALRGRYRGQTQSQYLAIDHGNGKDDGKAEAQSAADSSRASVSGTSGMGQAQSQSMYDPTCDDCVGRSTGIDTSKQKPLGDSHGGIPSGLPSGAIGTPGNNLPFDPTGGSRLTGNGGYGPNENGERYGPGSTGINGAYRPPGAGGTYGPNQIGNGAFNPALNGIPGIRVQGPNTGYGISPGGGYTGSSPGLGNQYLPPSTSTNYEPVKGSYGPRQNVPDGSINGYLPVPSGSNVYGPNPNSGSGHGIVSNIPGGGYGQNGNYGPNQNGNMPYGTNQVPSQGTKTPAHGGGPSGINTGLSWPSTGTFTRPGSQNTPNGPSSQGNANEPHNLLAPNAVYGPGGNRPYQTDGQSNYQTKPNNAYGTTGGYTPGSNGPFNPADIGTNNNANKYGQLNPNNQNGLQSPGTYSPTSTGQNHMPGVIGSQNGLQGPGGQYAPTNTDGQIPGVQYNPGSTVGHNGLPIPGGLYNSGIGGGPNSFPGTPGQYQPGSTDGHNIVTGPGVQYRPDIVSGQNGLQGPGSQYGGGQSRNQEPGGQYAPGNQNIMQRPSGQYGPDSAVGQGIQEPGNMYDSRPNNQSPQYGPGIGIGPNGLQGPTGQYRPGSSGGQNVVSGPGGQYGSSGTQYGPGGQNGIQGPSSQYGPGGQNGIQVPGTQYGPGGQNGIQVPGTEYGPGGQRGIQGPGTLYGPNGQNGIQSPGTQYGPGGQIGIQAPGTQYGPGEQNGIQSPGTKTGPEGQLGNQGPGTQYGPGGQYGIQGPGNQYVPGGQGGIQAPVSQYDNVQGGIRGPGTQYGPSGQSGLQGPGGQNGIQYPGGSYGTGVQSGLQKPGGVYGSGLNGIQNNGVPGTGDNYVPGGQSNVPGGYGISNRPFNLDGTGSQQGPYNPSLPGQNGPYGTNMVGQGSGSISPGTTGLGNTGNQNGQYGPSAGPYPNQYGNDLSNTGGGSGQYANVPYGYGNNQAGAPQNVVDPNSALLIDGDDSAAEASVSQASNGTTAIASSKGGNDKGRAQTHVQGAYTGGGSFSAQAEISGSETGSILTGSQSEGVMHSSNTQVQGSLKGGMADAQARGPGSTSSQAQIGFTPYTHGDASHDVQKSPFVGGGTAAAQSSGRTGSSQSQLRGTFKYGITYNGGAQAGASLDKDTVFSNRLPFDKIDVYDEKNKNINIETTEKSLDLLPTESTEIPLTTEHLSYDIPSTTPLDISANTEKIDPVYSENNDEYGSHSHSDHHKTEFAASTPTRSPPVESRRSFQSAYGTNSGDYDYTTDKEENQPEEYDMDDGFAPEGTDTSDNDQGEFANYNEFSRDSETHQSLHSPKRKGLEVRQTTGGNTQHIVLGALRDQNAEITQRNSERPDESKIYQPGERVPGTGGYTIPVGFTGSVKSVASKDKTYVVGSKDSPSQAQTVSLTPGSGKIKYTYPTTHTRNVSPKDLRSLYNVKPDDNNRYVSVSKSVTRDLDSDNNIRKQYSHTYYTKSSSCGYFTFTCNMVSGAEGKKKVCKPKIPTNPDGTPMRC
Summary
Uniprot
Pubmed
EMBL
Proteomes
PRIDE
ProteinModelPortal
Ontologies
GO
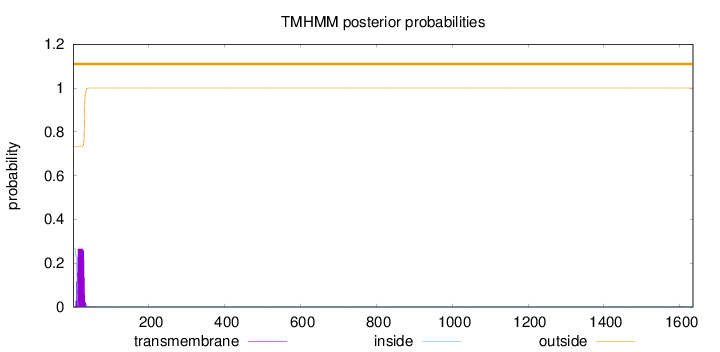
Topology
SignalP
Position: 1 - 30,
Likelihood: 0.742699
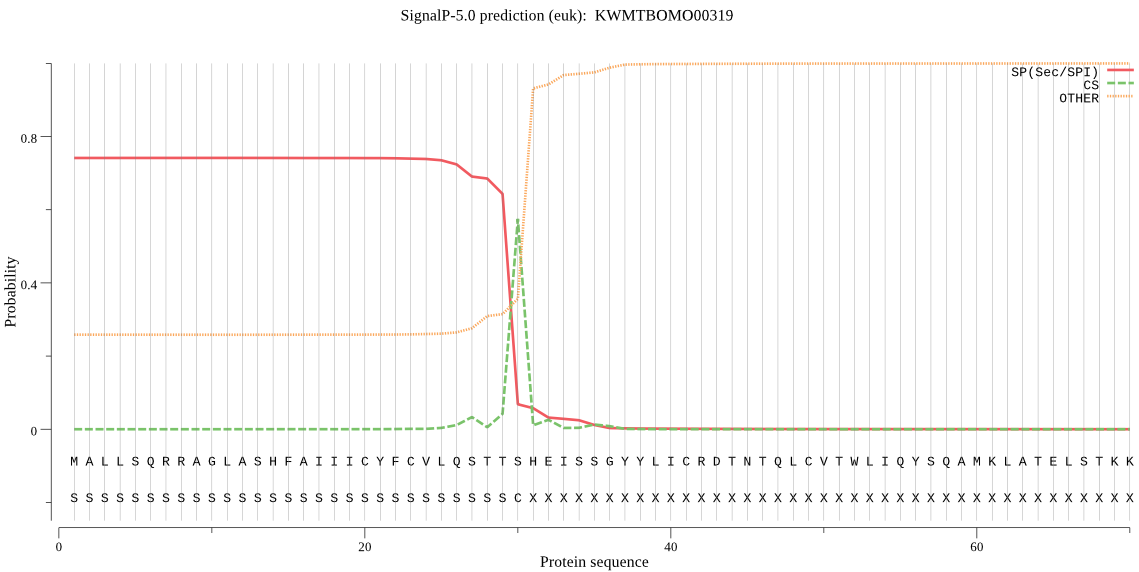
Length:
1635
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.39061
Exp number, first 60 AAs:
5.38433
Total prob of N-in:
0.26561
outside
1 - 1635
Population Genetic Test Statistics
Pi
1.753561
Theta
3.127984
Tajima's D
-1.150165
CLR
0
CSRT
0.104844757762112
Interpretation
Uncertain
