Gene
KWMTBOMO00314
Pre Gene Modal
BGIBMGA000537
Annotation
PREDICTED:_uncharacterized_protein_LOC101743416_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.626
Sequence
CDS
ATGAATGGAGAAATAAATGGAATTATGGATAAATCATTAGATTCACCTGCAGACCTATCTAACATACAAGATGACCATAAGAATTCAGATGCTAAAGATGATCATGATGACACTGAAAGTGATAGTAGTACCAATGTGAAAGGTAGTGTATTGATAACTCAAAGTATTACGGTAGATGATGGTTCTTCCATTGAAACCAAAACTAATAGTGATTCCGAGCAGTCTGACAGCAACAGTATTCATAATAATGATGATAAGATAATACTTGTACGTGAAGCTGGTACTGTAAAAATGGCCATCCAAGATGATAACTGTGTTACTGCAGAAAATGAATGTGGAAACAGCAATGACTCTTATACTGGTTCAGAAACCAGTTCACAGGCTAAGTCTATAGAGCAACTCAAAATCACCAGTATTGATATAAAATATAATTATACAGGAAAAGACACTTCAAATAGTTGTGACAATACACAAAACTCAGATAATACAGAGGAATCATCAGATGATTGTTCTGAGCCTAAAGCAGATGATGCAGTAAGCATCAACTCTGACTCTGGCGATGATGGATCTTCACAGGGAAGTGACAATAAACTTAAAAGAACAGAGGTAATCTTGATAGATAATGATAGGTCAGATACACAAAATGGGAATTCACATGATGCTCACAGTCCAGAAGTCCATGAAATAGAAAGTGATAATGATGAAACATTTACTGAACCAGTAGTAGAAAAACCAAAATCAAATACAAATAATAGCAAAACAGTTCTTAGAAGAAGCAGTAGAACAATAAAGAGGAGGAAATTCAGTGATGAAATAGAAAATGGTGATGACTCAGAAGTGGAAGAAATAGAAATGCTGGATCCTTTACGGCATAAAATGAGACCAATAGTCATCAATGATACTAAAACATTGGTTGAGATGGCAGCAAAACATACAAAGGCAAGTCAGAATGACAATAAGAAGAAAGAACCAACAGTTGTTATAATTGATACACATGCTAACAAGCAAACAATAAACAAAACACAACCTTCAATTAATTCTACAAATGCTCAAACTTTATATCAGAATATGATTGCTCGTGGAACTACAGTTACCCCTGTTAGTAGTAAAAGTAATGCAACAAACAATAATAGCTCCTCACAGACTCAAACAATTCTACCCTCACTTACAGATGATATGTATGTGGTTGAAGCACCATCTTTTATTGTACCATACGTGTACGAAAAACCCTCTTTGCAACCTTTTAGAGAATTTGTAGACATATTAGGTAAAGAGCTAGAAGAACAAAAAGCCAAAGAAGACAAAGAGCGATTAGAAAAAAATAACAAAGAGAAAGAGAAAAGAGCCAAGGAAATGCATGAAAAAATAACCAAAGGAGAAGAAGAAGTAGAGGAATCTGAAGAAGATGTGGATATGGAGAAAAAGAAGAAAACTGAAAAAAGAGAGCGACGAAGGCATGGGAATGATGATGATGATGCATCTTGGGATGGTGAGTCATCTTCAGAGTCTGAAGATGATGTAATTAGTGATGAAGAAGATAAAATTCTCAAAGATAAAGTTGATCCAATAGATGATATCAAGGATGTGACTGATATAGTAACTGACAAAACCGTTGGTAAATCTGAGAGCTACTTTGATTGTTCACTAGGCAAATTTTTTATGGATATTGGTTACAACCTTGTTCAAGAATTTGTACAAAGTGATTTGCTCAAACAGCAGAACCGTAAATTAAACAGGGAAAAGAAATCTGGCCATAGCACAAAGGCTACCGAATCATCAATAGCATCGCTCATGAAAAATTTGGAATTAAGCAAAGAAAATAATGCACCTTATGCTTATCTGCAGAATAAATGTCAATTTTGCACCTTTAAGACTGAGTCTACCCTTGTAATGGCCAATCATTTAGAAACACCTCACATGAAAAATAATTTGTACAAGTGTAACTTTTGTGAATTTGAAATTCGAAGCCCACATGATATTTTGAGGCATATGAAAGCTGAGCATAATATTGTAGGTAAATTAGAAAAAGCTCCATCTTATCACGAGTGTCCTAATTGCCCGTTTGAAGATAATGGTAAGGGTAAATTAGCTCGTCATCAAATACCTTGTGCTAAAAAATTTAAACCAGCTATCAATTTATCACCACCAATTGAATGGGAGCCCCCAGCAAAAATACCAAAGATAACAAAATCGCGTAGTAATGTGATGGGCCCGTATCAAAACCAGTTTAACCGCCATGTTGGTAATAATATTAACAGGCCAATGTCAGTAACAAATATGGCACCAGGAGGCAGTTTCCGCCCTCGAGGTAGAGCACCATTGTCAATGCCTCCTCGGGGCGCACCCATACCTGTTCATGGAGCTCCTATTATACGTGGAGGTGTTATGATTAGGCATAATTCTCCTGCTGCTTCTTCAAATTATCCGACAAAAAATAAGCCAGGGCAGCAGCCATCTATATCGATAACGCCCTTGCCACGGCAACCGCAACCTGCACAGGCAGTTCAGAATGTGCAGGCATCAAAAGGCTCGTTTGTGATTTGCGAGATCTGTGATGGTTACATAAAAGACCTAGAGCAATTGCGAAATCATATGCAATGGATTCACAAAGTTAAAATTCACCCAAAAATGATCTACAACAGACCACCACTGAACTGCCAGAAATGTCAATTTAGGTTTTTCACTGACCAAGGTTTGGAACGACATTTATTAGGCTCGCATGGTCTTGTTACTAGTTCTATGCAAGAAGCAGCAAATAAAGGCAAAGATGCAGGCCGATGTCCAGTTTGTGGCAGAGTATATCAATGGAAACTTCTTAATCATGTTGCTCGTGATCACAATCTAACATTAAAGCCAGCACATTTGTCTTATAAATGTACCGTGTGTACTGCCACATTCGGAATGTATAAACTGTTCGAGAACCATGTTTATTCAGCTCATAGTGTAGTTGCAAAACGCGTTATGGACAAAAACAAAAATAATACACCATCTGCCAAAGCTTCAGCAAACAACGACTCACTCTTGAAACCCTTGAAGATAAATGATGAGATTACTATTATCCCACAGCCAGTTAAAGCAAGTGCTGCTAATTGTGCTAAAGATAAATAA
Protein
MNGEINGIMDKSLDSPADLSNIQDDHKNSDAKDDHDDTESDSSTNVKGSVLITQSITVDDGSSIETKTNSDSEQSDSNSIHNNDDKIILVREAGTVKMAIQDDNCVTAENECGNSNDSYTGSETSSQAKSIEQLKITSIDIKYNYTGKDTSNSCDNTQNSDNTEESSDDCSEPKADDAVSINSDSGDDGSSQGSDNKLKRTEVILIDNDRSDTQNGNSHDAHSPEVHEIESDNDETFTEPVVEKPKSNTNNSKTVLRRSSRTIKRRKFSDEIENGDDSEVEEIEMLDPLRHKMRPIVINDTKTLVEMAAKHTKASQNDNKKKEPTVVIIDTHANKQTINKTQPSINSTNAQTLYQNMIARGTTVTPVSSKSNATNNNSSSQTQTILPSLTDDMYVVEAPSFIVPYVYEKPSLQPFREFVDILGKELEEQKAKEDKERLEKNNKEKEKRAKEMHEKITKGEEEVEESEEDVDMEKKKKTEKRERRRHGNDDDDASWDGESSSESEDDVISDEEDKILKDKVDPIDDIKDVTDIVTDKTVGKSESYFDCSLGKFFMDIGYNLVQEFVQSDLLKQQNRKLNREKKSGHSTKATESSIASLMKNLELSKENNAPYAYLQNKCQFCTFKTESTLVMANHLETPHMKNNLYKCNFCEFEIRSPHDILRHMKAEHNIVGKLEKAPSYHECPNCPFEDNGKGKLARHQIPCAKKFKPAINLSPPIEWEPPAKIPKITKSRSNVMGPYQNQFNRHVGNNINRPMSVTNMAPGGSFRPRGRAPLSMPPRGAPIPVHGAPIIRGGVMIRHNSPAASSNYPTKNKPGQQPSISITPLPRQPQPAQAVQNVQASKGSFVICEICDGYIKDLEQLRNHMQWIHKVKIHPKMIYNRPPLNCQKCQFRFFTDQGLERHLLGSHGLVTSSMQEAANKGKDAGRCPVCGRVYQWKLLNHVARDHNLTLKPAHLSYKCTVCTATFGMYKLFENHVYSAHSVVAKRVMDKNKNNTPSAKASANNDSLLKPLKINDEITIIPQPVKASAANCAKDK
Summary
Uniprot
Pubmed
Proteomes
PRIDE
Interpro
IPR013087
Znf_C2H2_type
ProteinModelPortal
Ontologies
GO
Topology
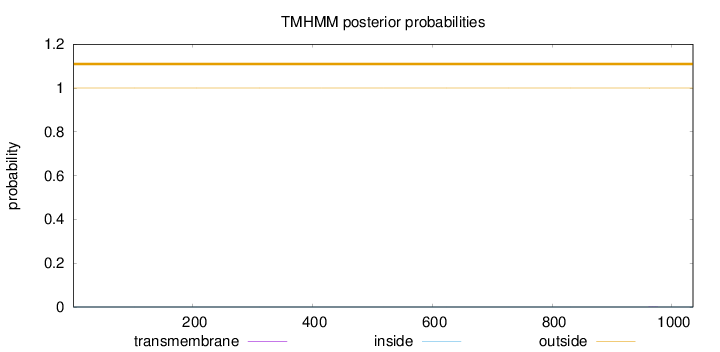
Length:
1035
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00391
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00000
outside
1 - 1035
Population Genetic Test Statistics
Pi
0.278944
Theta
3.609361
Tajima's D
-2.280116
CLR
2.438536
CSRT
0.0023998800059997
Interpretation
Possibly Positive selection
