Gene
KWMTBOMO00306
Pre Gene Modal
BGIBMGA000684
Annotation
PREDICTED:_insulin-like_growth_factor-binding_protein_complex_acid_labile_subunit_isoform_X1_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.107
Sequence
CDS
ATGTCATCTTTGGTCGTATATAATACGGAGGCCGCAGGGACAATCTGCCCTGATGAATGTACTTGTACTATGACATTTGATCTCGAAGAAGGCCATACAACGCAAGTGGCTTGTCAGCATGGACACATGAGGATAATCCCGGTCGACTACATGGACCGTGACGTGCATGTTGTTATTATAGCCGCTCCTCCGGATCGACCTAACTTTTTAACCATTGGACCCATATTTACTCAACCAGCCCCATTTGTAAACCTTCGCGAACTTCACATTGTTAATTCCAATGTACCGTCGATTGGGCAATTTTCCTTTTGGGGCCTGCAAAATTTAAGAGTTTTAAATTTGACACACAACAACCTTACGAGTGTAGTAGCTGATAATTTTAGAGGTCTCATAAATCTTACTGAACTTCATCTCGATCATAATCATATCGAACAGATGTCAAGCGAGACTTTCAGACACCTGACCTCATTAAAAACTCTAATATTGGCCAATAATCAAATATCGACACTCGTGTCGAGGTTGTTTAGAATGCTGGCGAAGTTGTCTATTTTGGATTTGAGTGACAACCCATTAACTGACTTGAACCCTGAAGTTTTTAAAGATATTCAACACTTGCGAATATTTAAATGTAGAAGATGTTTACTGAGACGCATAAACACACAAATTTATCATTTAGTACCTAGACTCGAGGAATTGGATTTGGGTGAAAATTTATTTAAATACTTAACATCAGATGAATTCATAAGCTTGAAGCGATTAAAAAAATTAAAATTGGACGGGAATCAATTGTCAGTAATCGTGGACAACATGTTTGGACGGAACCGGGAACTCCACGTACTAACATTATCACGCAACCGTCTAGCCCTCTTGGCGCCATTGGCCCTCACTAATCTAACCGCCCTCACACATTTGGATATAAGTCATAACAAAATTGACAGGTTCCATTTGCAAACTTTCGCTCCGGTTGTGGACTCCTTAAAAACAATAAATTTCAGTGGAAATAATTTACCACTAAACGAAATTGCTATCGTATTACAAATACTACCAGAGATTCATGGAGTAGGCTTAGCCAACTTGTCACTGCAAGAAATACCTCAAAATTTCTTTACATACAACGAGCATCTCGTGGCTTTGGACTTATCTTGGAATAAATTATCCAGGTTTCCCTTCAAACTTCTATCTAAGACAAAGTTTTTACAGCGATTAGATATATCGCACAACAAGTTACAAACCTTAAGCGAACAGGATTTGCAAAGATTGGAAGCTATAGCTGAAGTAAACTTAATGAAAAATAATTGGCATTGCGATCAATGTTCGGCGAGTAATATGCTTTTGTACATGACCACAACAGTCCTTAACAGTACAATACGAGTCCTCAAATGCCACTCCCCGTTAAGATTGCGAGGCGCAACTTTTGGTAGTCTGAGTTTTGAGCAATTGGATCCATGTCCGACGACACATGAAGCACAAATGAGTGTCATTGCGGGATTAATGCTGTTATGTGTAGCTGTCCTCTCAGCAGTAGCTGCGGCGCTTTGCTGTACACGACGTCGCGCCGCCCAATACTATACCAATGAAGAAAAGCGTAGAGAACACGATCATGAACATCCAGAAGAACTGCTTGGACATACGGAACTTGGCAGCGTCGCTACTATCCACGCGCCGTACCAACTCGTCTCTAAGGACAGCTAA
Protein
MSSLVVYNTEAAGTICPDECTCTMTFDLEEGHTTQVACQHGHMRIIPVDYMDRDVHVVIIAAPPDRPNFLTIGPIFTQPAPFVNLRELHIVNSNVPSIGQFSFWGLQNLRVLNLTHNNLTSVVADNFRGLINLTELHLDHNHIEQMSSETFRHLTSLKTLILANNQISTLVSRLFRMLAKLSILDLSDNPLTDLNPEVFKDIQHLRIFKCRRCLLRRINTQIYHLVPRLEELDLGENLFKYLTSDEFISLKRLKKLKLDGNQLSVIVDNMFGRNRELHVLTLSRNRLALLAPLALTNLTALTHLDISHNKIDRFHLQTFAPVVDSLKTINFSGNNLPLNEIAIVLQILPEIHGVGLANLSLQEIPQNFFTYNEHLVALDLSWNKLSRFPFKLLSKTKFLQRLDISHNKLQTLSEQDLQRLEAIAEVNLMKNNWHCDQCSASNMLLYMTTTVLNSTIRVLKCHSPLRLRGATFGSLSFEQLDPCPTTHEAQMSVIAGLMLLCVAVLSAVAAALCCTRRRAAQYYTNEEKRREHDHEHPEELLGHTELGSVATIHAPYQLVSKDS
Summary
Uniprot
H9ITV4
A0A2A4J7G7
A0A194QVC6
A0A2H1WC35
A0A194PTM7
A0A212ETF3
+ More
A0A0L7LKI9 A0A2P8XRJ1 A0A067QIL7 A0A1B6CGE8 A0A1B6M847 D6WMZ1 A0A2R7W3J2 A0A1S3CYG3 A0A1J1HVX9 Q16P50 A0A1W4WD78 A0A1I8MYU9 W8BWZ3 A0A182REW8 A0A182WGL1 A0A1I8PV42 A0A2P6L4Z1 N6UNP4 A0A1A9WMF3 A0A226ES49 A0A1Y1KFD8 A0A182M6A0 A0A1B0EYS2 A0A087SZ97 A0A182F2W7 A0A0L0BY75 A0A1B0AGS2 A0A0A1XCU3 B4JCA8 A0A1A9YFQ8 W5JC04 A0A336M357 A0A336LR43 A0A182YPT0 A0A0T6BA38 T1JDL4 A0A0K8UAG2 A0A0K8UDI3 F0JAM7 A0A084WG87 A0A182VHT2 A0A182JGD4 A0A3B0JD60 Q7Q1I6 A0A182XNA9 A0A182NBT4 A0A182KTZ6 A0A182U0Z6 B3MMF1 B4MYU0 B4Q8D7 B3NME1 A0A336LUR9 Q9VJA9 A0A1B0APZ5 A0A0R1DNT0 A0A182HRV3 B4P8N5 Q29LC8 A0A1B0G273 B4GQ71 B4I5E1 A0A1W4VHG8 D6WMY6 A0A0Q9WN82 B4KEF9 B4LSW3 A0A386H7K0 A0A1W4WJZ7 A0A131YZV6 A0A0Q9XIC9 X2J6K6 A0A182PUX8 A0A226DE00 A0A182KDI6 A0A0P4WAM5 A0A0P5DDD8 A0A0P5DF35 A0A0P5Y4R8 A0A0P4YYW4 A0A0P6GKW4 A0A0P4YDI6 A0A0P5TM99 A0A0P6C180 A0A0P5VLI3 A0A182Q1C3 A0A0P5VP45 A0A087UKM2 A0A0N8D3R3 A0A1B0CB91
A0A0L7LKI9 A0A2P8XRJ1 A0A067QIL7 A0A1B6CGE8 A0A1B6M847 D6WMZ1 A0A2R7W3J2 A0A1S3CYG3 A0A1J1HVX9 Q16P50 A0A1W4WD78 A0A1I8MYU9 W8BWZ3 A0A182REW8 A0A182WGL1 A0A1I8PV42 A0A2P6L4Z1 N6UNP4 A0A1A9WMF3 A0A226ES49 A0A1Y1KFD8 A0A182M6A0 A0A1B0EYS2 A0A087SZ97 A0A182F2W7 A0A0L0BY75 A0A1B0AGS2 A0A0A1XCU3 B4JCA8 A0A1A9YFQ8 W5JC04 A0A336M357 A0A336LR43 A0A182YPT0 A0A0T6BA38 T1JDL4 A0A0K8UAG2 A0A0K8UDI3 F0JAM7 A0A084WG87 A0A182VHT2 A0A182JGD4 A0A3B0JD60 Q7Q1I6 A0A182XNA9 A0A182NBT4 A0A182KTZ6 A0A182U0Z6 B3MMF1 B4MYU0 B4Q8D7 B3NME1 A0A336LUR9 Q9VJA9 A0A1B0APZ5 A0A0R1DNT0 A0A182HRV3 B4P8N5 Q29LC8 A0A1B0G273 B4GQ71 B4I5E1 A0A1W4VHG8 D6WMY6 A0A0Q9WN82 B4KEF9 B4LSW3 A0A386H7K0 A0A1W4WJZ7 A0A131YZV6 A0A0Q9XIC9 X2J6K6 A0A182PUX8 A0A226DE00 A0A182KDI6 A0A0P4WAM5 A0A0P5DDD8 A0A0P5DF35 A0A0P5Y4R8 A0A0P4YYW4 A0A0P6GKW4 A0A0P4YDI6 A0A0P5TM99 A0A0P6C180 A0A0P5VLI3 A0A182Q1C3 A0A0P5VP45 A0A087UKM2 A0A0N8D3R3 A0A1B0CB91
Pubmed
19121390
26354079
22118469
26227816
29403074
24845553
+ More
18362917 19820115 17510324 25315136 24495485 23537049 28004739 26108605 25830018 17994087 20920257 23761445 25244985 24438588 12364791 20966253 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243 18057021 26830274
18362917 19820115 17510324 25315136 24495485 23537049 28004739 26108605 25830018 17994087 20920257 23761445 25244985 24438588 12364791 20966253 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 23185243 18057021 26830274
EMBL
BABH01007044
NWSH01002890
PCG67362.1
KQ461108
KPJ09274.1
ODYU01007655
+ More
SOQ50623.1 KQ459594 KPI96109.1 AGBW02012593 OWR44782.1 JTDY01000765 KOB75962.1 PYGN01001473 PSN34628.1 KK853309 KDR08672.1 GEDC01024786 JAS12512.1 GEBQ01007874 JAT32103.1 KQ971343 EFA03262.2 KK854293 PTY14284.1 CVRI01000024 CRK92173.1 CH477794 EAT36134.1 GAMC01005152 JAC01404.1 MWRG01001786 PRD33658.1 APGK01024659 KB740558 KB632357 ENN80352.1 ERL93465.1 LNIX01000002 OXA59416.1 GEZM01089477 JAV57487.1 AXCM01004563 AJVK01005576 AJVK01005577 KK112645 KFM58186.1 JRES01001166 KNC24921.1 GBXI01005526 JAD08766.1 CH916368 EDW04141.1 ADMH02001647 ETN61576.1 UFQT01000121 SSX20428.1 SSX20430.1 LJIG01002738 KRT84202.1 JH432107 GDHF01028690 GDHF01026135 JAI23624.1 JAI26179.1 GDHF01027678 JAI24636.1 BT126069 ADY17768.1 ATLV01023473 KE525343 KFB49231.1 OUUW01000004 SPP79995.1 AAAB01008980 EAA14468.4 CH902620 EDV30897.1 CH963913 EDW77279.2 CM000361 CM002910 EDX05347.1 KMY90762.1 CH954179 EDV54812.1 SSX20429.1 AE014134 BT004502 AAF53643.2 AAO42666.1 JXJN01001618 CM000158 KRJ98954.1 APCN01001755 APCN01001756 EDW90143.1 CH379060 EAL34117.1 KRT04480.1 KRT04481.1 KRT04482.1 CCAG010011277 CCAG010011278 CH479187 EDW39743.1 CH480822 EDW55597.1 EFA04323.2 CH940649 KRF81909.1 CH933807 EDW12927.2 EDW64874.1 KRF81907.1 KRF81908.1 KRF81910.1 MG452691 AYD41578.1 GEDV01004379 JAP84178.1 KRG03496.1 AHN54538.1 LNIX01000022 OXA43350.1 GDRN01073811 JAI63330.1 GDIP01173966 JAJ49436.1 GDIP01157237 JAJ66165.1 GDIP01062996 JAM40719.1 GDIP01220122 JAJ03280.1 GDIQ01032062 GDIQ01001434 JAN62675.1 GDIP01230068 GDIP01187724 GDIP01125419 GDIP01124372 JAI93333.1 GDIP01124373 LRGB01000005 JAL79341.1 KZS22027.1 GDIP01006268 JAM97447.1 GDIP01113033 JAL90681.1 AXCN02000690 GDIP01097379 JAM06336.1 KK120269 KFM77911.1 GDIP01071743 JAM31972.1 AJWK01004899
SOQ50623.1 KQ459594 KPI96109.1 AGBW02012593 OWR44782.1 JTDY01000765 KOB75962.1 PYGN01001473 PSN34628.1 KK853309 KDR08672.1 GEDC01024786 JAS12512.1 GEBQ01007874 JAT32103.1 KQ971343 EFA03262.2 KK854293 PTY14284.1 CVRI01000024 CRK92173.1 CH477794 EAT36134.1 GAMC01005152 JAC01404.1 MWRG01001786 PRD33658.1 APGK01024659 KB740558 KB632357 ENN80352.1 ERL93465.1 LNIX01000002 OXA59416.1 GEZM01089477 JAV57487.1 AXCM01004563 AJVK01005576 AJVK01005577 KK112645 KFM58186.1 JRES01001166 KNC24921.1 GBXI01005526 JAD08766.1 CH916368 EDW04141.1 ADMH02001647 ETN61576.1 UFQT01000121 SSX20428.1 SSX20430.1 LJIG01002738 KRT84202.1 JH432107 GDHF01028690 GDHF01026135 JAI23624.1 JAI26179.1 GDHF01027678 JAI24636.1 BT126069 ADY17768.1 ATLV01023473 KE525343 KFB49231.1 OUUW01000004 SPP79995.1 AAAB01008980 EAA14468.4 CH902620 EDV30897.1 CH963913 EDW77279.2 CM000361 CM002910 EDX05347.1 KMY90762.1 CH954179 EDV54812.1 SSX20429.1 AE014134 BT004502 AAF53643.2 AAO42666.1 JXJN01001618 CM000158 KRJ98954.1 APCN01001755 APCN01001756 EDW90143.1 CH379060 EAL34117.1 KRT04480.1 KRT04481.1 KRT04482.1 CCAG010011277 CCAG010011278 CH479187 EDW39743.1 CH480822 EDW55597.1 EFA04323.2 CH940649 KRF81909.1 CH933807 EDW12927.2 EDW64874.1 KRF81907.1 KRF81908.1 KRF81910.1 MG452691 AYD41578.1 GEDV01004379 JAP84178.1 KRG03496.1 AHN54538.1 LNIX01000022 OXA43350.1 GDRN01073811 JAI63330.1 GDIP01173966 JAJ49436.1 GDIP01157237 JAJ66165.1 GDIP01062996 JAM40719.1 GDIP01220122 JAJ03280.1 GDIQ01032062 GDIQ01001434 JAN62675.1 GDIP01230068 GDIP01187724 GDIP01125419 GDIP01124372 JAI93333.1 GDIP01124373 LRGB01000005 JAL79341.1 KZS22027.1 GDIP01006268 JAM97447.1 GDIP01113033 JAL90681.1 AXCN02000690 GDIP01097379 JAM06336.1 KK120269 KFM77911.1 GDIP01071743 JAM31972.1 AJWK01004899
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000037510
+ More
UP000245037 UP000027135 UP000007266 UP000079169 UP000183832 UP000008820 UP000192223 UP000095301 UP000075900 UP000075920 UP000095300 UP000019118 UP000030742 UP000091820 UP000198287 UP000075883 UP000092462 UP000054359 UP000069272 UP000037069 UP000092445 UP000001070 UP000092443 UP000000673 UP000076408 UP000030765 UP000075903 UP000075880 UP000268350 UP000007062 UP000076407 UP000075884 UP000075882 UP000075902 UP000007801 UP000007798 UP000000304 UP000008711 UP000000803 UP000092460 UP000002282 UP000075840 UP000001819 UP000092444 UP000008744 UP000001292 UP000192221 UP000008792 UP000009192 UP000075885 UP000075881 UP000076858 UP000075886 UP000092461
UP000245037 UP000027135 UP000007266 UP000079169 UP000183832 UP000008820 UP000192223 UP000095301 UP000075900 UP000075920 UP000095300 UP000019118 UP000030742 UP000091820 UP000198287 UP000075883 UP000092462 UP000054359 UP000069272 UP000037069 UP000092445 UP000001070 UP000092443 UP000000673 UP000076408 UP000030765 UP000075903 UP000075880 UP000268350 UP000007062 UP000076407 UP000075884 UP000075882 UP000075902 UP000007801 UP000007798 UP000000304 UP000008711 UP000000803 UP000092460 UP000002282 UP000075840 UP000001819 UP000092444 UP000008744 UP000001292 UP000192221 UP000008792 UP000009192 UP000075885 UP000075881 UP000076858 UP000075886 UP000092461
Interpro
Gene 3D
ProteinModelPortal
H9ITV4
A0A2A4J7G7
A0A194QVC6
A0A2H1WC35
A0A194PTM7
A0A212ETF3
+ More
A0A0L7LKI9 A0A2P8XRJ1 A0A067QIL7 A0A1B6CGE8 A0A1B6M847 D6WMZ1 A0A2R7W3J2 A0A1S3CYG3 A0A1J1HVX9 Q16P50 A0A1W4WD78 A0A1I8MYU9 W8BWZ3 A0A182REW8 A0A182WGL1 A0A1I8PV42 A0A2P6L4Z1 N6UNP4 A0A1A9WMF3 A0A226ES49 A0A1Y1KFD8 A0A182M6A0 A0A1B0EYS2 A0A087SZ97 A0A182F2W7 A0A0L0BY75 A0A1B0AGS2 A0A0A1XCU3 B4JCA8 A0A1A9YFQ8 W5JC04 A0A336M357 A0A336LR43 A0A182YPT0 A0A0T6BA38 T1JDL4 A0A0K8UAG2 A0A0K8UDI3 F0JAM7 A0A084WG87 A0A182VHT2 A0A182JGD4 A0A3B0JD60 Q7Q1I6 A0A182XNA9 A0A182NBT4 A0A182KTZ6 A0A182U0Z6 B3MMF1 B4MYU0 B4Q8D7 B3NME1 A0A336LUR9 Q9VJA9 A0A1B0APZ5 A0A0R1DNT0 A0A182HRV3 B4P8N5 Q29LC8 A0A1B0G273 B4GQ71 B4I5E1 A0A1W4VHG8 D6WMY6 A0A0Q9WN82 B4KEF9 B4LSW3 A0A386H7K0 A0A1W4WJZ7 A0A131YZV6 A0A0Q9XIC9 X2J6K6 A0A182PUX8 A0A226DE00 A0A182KDI6 A0A0P4WAM5 A0A0P5DDD8 A0A0P5DF35 A0A0P5Y4R8 A0A0P4YYW4 A0A0P6GKW4 A0A0P4YDI6 A0A0P5TM99 A0A0P6C180 A0A0P5VLI3 A0A182Q1C3 A0A0P5VP45 A0A087UKM2 A0A0N8D3R3 A0A1B0CB91
A0A0L7LKI9 A0A2P8XRJ1 A0A067QIL7 A0A1B6CGE8 A0A1B6M847 D6WMZ1 A0A2R7W3J2 A0A1S3CYG3 A0A1J1HVX9 Q16P50 A0A1W4WD78 A0A1I8MYU9 W8BWZ3 A0A182REW8 A0A182WGL1 A0A1I8PV42 A0A2P6L4Z1 N6UNP4 A0A1A9WMF3 A0A226ES49 A0A1Y1KFD8 A0A182M6A0 A0A1B0EYS2 A0A087SZ97 A0A182F2W7 A0A0L0BY75 A0A1B0AGS2 A0A0A1XCU3 B4JCA8 A0A1A9YFQ8 W5JC04 A0A336M357 A0A336LR43 A0A182YPT0 A0A0T6BA38 T1JDL4 A0A0K8UAG2 A0A0K8UDI3 F0JAM7 A0A084WG87 A0A182VHT2 A0A182JGD4 A0A3B0JD60 Q7Q1I6 A0A182XNA9 A0A182NBT4 A0A182KTZ6 A0A182U0Z6 B3MMF1 B4MYU0 B4Q8D7 B3NME1 A0A336LUR9 Q9VJA9 A0A1B0APZ5 A0A0R1DNT0 A0A182HRV3 B4P8N5 Q29LC8 A0A1B0G273 B4GQ71 B4I5E1 A0A1W4VHG8 D6WMY6 A0A0Q9WN82 B4KEF9 B4LSW3 A0A386H7K0 A0A1W4WJZ7 A0A131YZV6 A0A0Q9XIC9 X2J6K6 A0A182PUX8 A0A226DE00 A0A182KDI6 A0A0P4WAM5 A0A0P5DDD8 A0A0P5DF35 A0A0P5Y4R8 A0A0P4YYW4 A0A0P6GKW4 A0A0P4YDI6 A0A0P5TM99 A0A0P6C180 A0A0P5VLI3 A0A182Q1C3 A0A0P5VP45 A0A087UKM2 A0A0N8D3R3 A0A1B0CB91
PDB
5Z8Y
E-value=2.22068e-22,
Score=263
Ontologies
GO
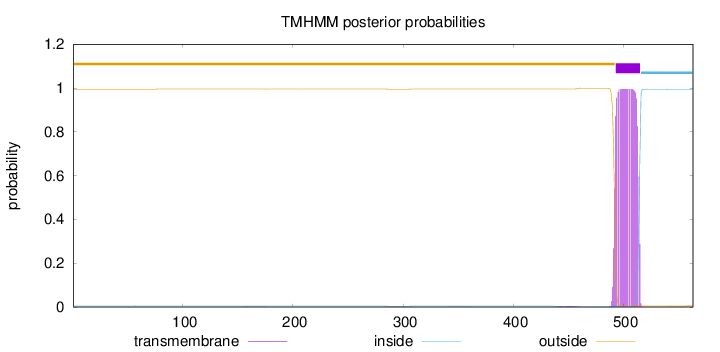
Topology
Length:
563
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.46671
Exp number, first 60 AAs:
0.00034
Total prob of N-in:
0.00496
outside
1 - 492
TMhelix
493 - 515
inside
516 - 563
Population Genetic Test Statistics
Pi
5.678825
Theta
5.572827
Tajima's D
-0.506951
CLR
0.023838
CSRT
0.241437928103595
Interpretation
Uncertain
