Pre Gene Modal
BGIBMGA000540
Annotation
PREDICTED:_ecotropic_viral_integration_site_5_ortholog_isoform_X5_[Bombyx_mori]
Full name
Ecotropic viral integration site 5 ortholog
Location in the cell
Cytoplasmic Reliability : 1.964 Nuclear Reliability : 1.344
Sequence
CDS
ATGACCGATCGGGCTTCAACTGAAAAATCTCCGGAGCTCAATCCAAACGCAGACTGCCGAATCCCGACAAGTGAAACTATCCCGACGCCCGACCGGGCGCTTCTCGCCAAGCTGGAGGAAGAAAACCGTCGTATCGAGGCTGATGCGAAGAACGCCTCGCTCACTACCGTCCACAGTCGGAAGAGCTCCGACACATCACTGATATCCCTCGCTTCCGCCACAAGCTCAAGTCATGAGGTCGAAACGCGCCCAACTCCTGGGACTAATGGAGAAGAGGATCTTTGGAGCCTCTGGGGTCGCCTCGTCAGCAACTGGGAATCGGAATGGAAACGTCGAAACCAATGGGTCCGCGACCTTGTTCGACAGGGTGTCCCACATCATTTCAGAGGCATCGTATGGCAGCTACTGGCAGGCGTAGACACGTCGCCCGAAAAGAAGTTATACGCCTCATATATTAAGGCGAAATCAGCCTGCGAGAAGGTCATACGTCGTGACATCGCTCGCACATATCCCGAACACGATTTCTTCAAAGAGAAGGATGGCCTCGGCCAGGAGTCTTTATTCAATGTGATGAAGGCGTATTCTTTGCACGATCGAGAAGTCGGCTACTGTCAAGGCTCAGGATTTATCGTCGGCTTATTACTAATGCAGATGCCGGAAGAGGAAGCGTTCGCTGTTTTAGTTAAAATAATGCAACAACATCGCATGCGGGACATGTTCAAACCGAGCATGGCGGAACTGGGACTCTGCATGTTTCAGCTGGAGAACTTGGTACAGGAACTGTTGCCTGACTTGCATGTTCACTTCCAATCTCAGAGCTTCAGCACGTCTCTGTACGCTAGTAGTTGGTTCCTGACTTTGTTTACAACAACACTATCACTTCCGCTCGCTTGCCGTATAATGGATGTGTTTCTCTCTGAAGGAATCGAAATTGTTTTCAAAGTCGCCCTAGCGTTGCTTACTTTAGGAAAAAACGATCTCTTATCGCTCGATATGGAGAGCATTTTGAAGTACATACAGAAAGAACTGCCCTTGAAAGCTGAAGCGGATCAAGCCGCTTTCATGAACTTGGCGTATTCAGTCAAAGTTAATCCGAAAAAGATGAAGAAACTCGAAAAGGAGTACACGGTTATAAAAACAAAGGAGCAAGGTGATATTGCCGTGCTCAGGTGTCTTCGTCAAGAGAACCGTCTCCTCAAGCAACGCGTAGAACTGCTGGAGAAGGAAAGTTCCGCTCTAGCAGAGCGTCTGGTCCGAGGTCAAGTGGACCGCGCTGAAGGAGAAGAAGAAACCTTTGCATTGGATCGAGAGGTCCAAGCGTTGCGTCGAGCTAACGTAGACATGCAGCAGCGGCTGGCCGTTGCTTTAGATGAGATAAGATCTCTCGAGATGACTATCGCCGAGAATAACTCGAGGCAGTCGTCGCTGGAGGGCACCGAAAGCGCCAGCGGTCAGAAACACGAGGAACTAGCGCGCTGCCTGCAGAGGGAGCTAGTGCGGGCGCGTCTACACGCAGCGGAACGCGAGGCCGCCGAACGCGAACTGACGGCTCGCATCGACGAACTCGAGAACGAGAACAAGAGCCTCAGGAGACAGCGCGTCGACAACAATGTCGCACACTTACAGGACGAGCTGATAGCCGTAAAGTTGCGTGAAGCTGAAGCTAATCTCTCGTTGAAGGACTTAAGGCAGCGGGTCTCGGAGCTGTCAGAATCGTGGCAACGACATTTGCAGGAGCACAAGCAGGAAGCACCCGCTGCTCCAGTTCAGTCGAATGTAGTATCAGACATCATGGCGACCCCGAAGAAACTGCTGCGAGCTTGGGAAGGGAGGTCGGCTGATATGCTCAAACTGGACGAGGAACTCATGACCACCAGGATCAGGGAGGTCGAGGCCTTGACCGAATTAAAGGAGTTAAGACTGAAGGTAATGGAACTAGAGACTCAAGTTCAAGTGTCGACAAATCAGCTCCGAAGACAGGACGAGGAAGCTCGGCTGTTACGCGAGAATCTAGATGCGGCCCTCCAACGTGAACGAGCTCTCCAAACGAGACAGAGAGAGTTCCAGCACAAATACGCTGATTTAGAGAGCAAGGCCAAGTATGAATCGATGCAAGCTAACATTCGAAACATGGAAGACGCGCAAAGAATAGCCGAACTGGAGACAGAAGTGTCTGAATATAAATTAAAGAATGAAGTCATGGCAACCGAAGGTGAACTAAGGAACAATAATATGGACGACTCCGACAGAGTACGTGAATTGCAAGAGCAAGTCGCCGAGCTAAAAGCGGAGATCATGAGGTTAGAGGCGTGGAAATCTCGGGCTCTCGGTCACACGGAGCTCAGCCGCGCCATATCCCTGGAGGACGATTTGGAAGAAGACGAGAAACTCAAGTTGACACTGCGCCGAGAATCTTCTACATCATTAGAATTTACAACAACCTCACGCAAACCTTCGTAG
Protein
MTDRASTEKSPELNPNADCRIPTSETIPTPDRALLAKLEEENRRIEADAKNASLTTVHSRKSSDTSLISLASATSSSHEVETRPTPGTNGEEDLWSLWGRLVSNWESEWKRRNQWVRDLVRQGVPHHFRGIVWQLLAGVDTSPEKKLYASYIKAKSACEKVIRRDIARTYPEHDFFKEKDGLGQESLFNVMKAYSLHDREVGYCQGSGFIVGLLLMQMPEEEAFAVLVKIMQQHRMRDMFKPSMAELGLCMFQLENLVQELLPDLHVHFQSQSFSTSLYASSWFLTLFTTTLSLPLACRIMDVFLSEGIEIVFKVALALLTLGKNDLLSLDMESILKYIQKELPLKAEADQAAFMNLAYSVKVNPKKMKKLEKEYTVIKTKEQGDIAVLRCLRQENRLLKQRVELLEKESSALAERLVRGQVDRAEGEEETFALDREVQALRRANVDMQQRLAVALDEIRSLEMTIAENNSRQSSLEGTESASGQKHEELARCLQRELVRARLHAAEREAAERELTARIDELENENKSLRRQRVDNNVAHLQDELIAVKLREAEANLSLKDLRQRVSELSESWQRHLQEHKQEAPAAPVQSNVVSDIMATPKKLLRAWEGRSADMLKLDEELMTTRIREVEALTELKELRLKVMELETQVQVSTNQLRRQDEEARLLRENLDAALQRERALQTRQREFQHKYADLESKAKYESMQANIRNMEDAQRIAELETEVSEYKLKNEVMATEGELRNNNMDDSDRVRELQEQVAELKAEIMRLEAWKSRALGHTELSRAISLEDDLEEDEKLKLTLRRESSTSLEFTTTSRKPS
Summary
Description
Functions as a GTPase-activating protein (GAP). During border cell migration in the ovary, acts as a GAP for Rab11 and is necessary for the maintenance of active receptor tyrosine kinases at the leading edge.
Subunit
Interacts with Rab11.
Keywords
Alternative splicing
Coiled coil
Complete proteome
Cytoplasm
Endosome
GTPase activation
Phosphoprotein
Reference proteome
Feature
chain Ecotropic viral integration site 5 ortholog
splice variant In isoform C and isoform D.
splice variant In isoform C and isoform D.
Uniprot
H9ITG0
A0A2A4J4U2
A0A2A4J634
A0A1E1W9Y6
A0A212ETK5
A0A2H1WCQ3
+ More
A0A2A4J422 A0A194PY85 A0A194QUR6 A0A2J7RJB0 A0A158NCK4 A0A195BYH3 A0A195F9B5 A0A067RH37 E2ALY0 A0A2J7RJA0 A0A3L8DC59 A0A1B6MKB5 A0A0M8ZPT1 A0A2A3EIJ2 A0A151X684 A0A0L7R8G4 A0A232F3K9 E2BM66 A0A1B6C0N9 K7J5B5 A0A1B6KW95 A0A026WJY9 D6WFG7 A0A224X7U8 A0A1B6EEN8 X1WJP3 E9IMA2 A0A1Y1KV77 A0A182UTN2 A0A182TJF7 A0A182XIC9 A0A182LFG3 N6TD12 A0A0C9RTX4 U4U569 A0A146KNV6 A0A1J1IUI5 Q7QBX8 A0A146L1Y0 A0A1J1IYV9 A0A182HTB4 F5HK77 A0A0V0GC26 A0A2S2NMS2 A0A146LNM6 A0A2M4BEC2 W5JEK1 A0A2M3Z0I4 A0A182F8N7 A0A1W4XH45 A0A1W4XSP3 A0A084VJR8 A0A182RCW4 A0A182Q3D9 A0A1B6MEE3 A0A182Y798 A0A182K833 A0A1B6EDA2 A0A182INU1 F4WX67 A0A182N4N1 A0A182VQ95 T1HVV6 A0A1W4XSK6 A0A182PA27 A0A1W4XRP5 A0A0Q9WZU9 A0A2M4A727 A0A0Q9WP52 U5EYP4 A0A0Q9W711 A0A0K8TRU0 B4L4A9 A0A2M4A7R0 A0A1B0CM39 A0A2M4BEZ8 B4MCX5 Q9VYY9-4 A0A0Q9WRJ7 A0A0Q5TIT0 A0A1B6CY08 A0A0R1EBB0 E0W1N8 Q9VYY9 A0A1L8DVN3 B4JIW9 B3NUD4 A0A023EXU8 Q16J34 A0A3B0K982 B4PYA9 A0A1W4UNL1 A0A1Q3EZ55 B4N1T8
A0A2A4J422 A0A194PY85 A0A194QUR6 A0A2J7RJB0 A0A158NCK4 A0A195BYH3 A0A195F9B5 A0A067RH37 E2ALY0 A0A2J7RJA0 A0A3L8DC59 A0A1B6MKB5 A0A0M8ZPT1 A0A2A3EIJ2 A0A151X684 A0A0L7R8G4 A0A232F3K9 E2BM66 A0A1B6C0N9 K7J5B5 A0A1B6KW95 A0A026WJY9 D6WFG7 A0A224X7U8 A0A1B6EEN8 X1WJP3 E9IMA2 A0A1Y1KV77 A0A182UTN2 A0A182TJF7 A0A182XIC9 A0A182LFG3 N6TD12 A0A0C9RTX4 U4U569 A0A146KNV6 A0A1J1IUI5 Q7QBX8 A0A146L1Y0 A0A1J1IYV9 A0A182HTB4 F5HK77 A0A0V0GC26 A0A2S2NMS2 A0A146LNM6 A0A2M4BEC2 W5JEK1 A0A2M3Z0I4 A0A182F8N7 A0A1W4XH45 A0A1W4XSP3 A0A084VJR8 A0A182RCW4 A0A182Q3D9 A0A1B6MEE3 A0A182Y798 A0A182K833 A0A1B6EDA2 A0A182INU1 F4WX67 A0A182N4N1 A0A182VQ95 T1HVV6 A0A1W4XSK6 A0A182PA27 A0A1W4XRP5 A0A0Q9WZU9 A0A2M4A727 A0A0Q9WP52 U5EYP4 A0A0Q9W711 A0A0K8TRU0 B4L4A9 A0A2M4A7R0 A0A1B0CM39 A0A2M4BEZ8 B4MCX5 Q9VYY9-4 A0A0Q9WRJ7 A0A0Q5TIT0 A0A1B6CY08 A0A0R1EBB0 E0W1N8 Q9VYY9 A0A1L8DVN3 B4JIW9 B3NUD4 A0A023EXU8 Q16J34 A0A3B0K982 B4PYA9 A0A1W4UNL1 A0A1Q3EZ55 B4N1T8
Pubmed
19121390
22118469
26354079
21347285
24845553
20798317
+ More
30249741 28648823 20075255 24508170 18362917 19820115 21282665 28004739 20966253 23537049 26823975 12364791 20920257 23761445 24438588 25244985 21719571 17994087 18057021 26369729 10731132 12537572 12537569 17372656 18327897 22778279 17550304 20566863 24945155 17510324
30249741 28648823 20075255 24508170 18362917 19820115 21282665 28004739 20966253 23537049 26823975 12364791 20920257 23761445 24438588 25244985 21719571 17994087 18057021 26369729 10731132 12537572 12537569 17372656 18327897 22778279 17550304 20566863 24945155 17510324
EMBL
BABH01007029
BABH01007030
NWSH01003131
PCG66899.1
PCG66900.1
GDQN01007299
+ More
JAT83755.1 AGBW02012593 OWR44781.1 ODYU01007655 SOQ50622.1 PCG66897.1 KQ459594 KPI96110.1 KQ461108 KPJ09273.1 NEVH01002994 PNF40912.1 ADTU01011953 KQ976396 KYM92988.1 KQ981727 KYN36971.1 KK852685 KDR18482.1 GL440682 EFN65572.1 PNF40910.1 QOIP01000010 RLU18055.1 GEBQ01024317 GEBQ01003623 JAT15660.1 JAT36354.1 KQ435922 KOX68640.1 KZ288232 PBC31530.1 KQ982490 KYQ55788.1 KQ414632 KOC67123.1 NNAY01001051 OXU25331.1 GL449158 EFN83192.1 GEDC01030221 JAS07077.1 AAZX01000334 GEBQ01026154 GEBQ01024264 JAT13823.1 JAT15713.1 KK107167 EZA56367.1 KQ971327 EFA01267.1 GFTR01007979 JAW08447.1 GEDC01000906 JAS36392.1 ABLF02040575 ABLF02040576 GL764129 EFZ18387.1 GEZM01076826 JAV63615.1 APGK01043126 APGK01043127 APGK01043128 APGK01043129 KB741011 ENN75573.1 GBYB01011036 JAG80803.1 KB631957 ERL87468.1 GDHC01020521 JAP98107.1 CVRI01000059 CRL03768.1 AAAB01008859 EAA08217.5 GDHC01016241 JAQ02388.1 CRL03769.1 APCN01000435 EGK96918.1 GECL01001202 JAP04922.1 GGMR01005871 MBY18490.1 GDHC01010247 JAQ08382.1 GGFJ01002226 MBW51367.1 ADMH02001618 ETN61773.1 GGFM01001288 MBW22039.1 ATLV01013859 KE524905 KFB38212.1 AXCN02000052 GEBQ01031355 GEBQ01005695 JAT08622.1 JAT34282.1 GEDC01001382 JAS35916.1 GL888417 EGI61310.1 ACPB03020531 CH933810 KRF94026.1 GGFK01003230 MBW36551.1 KRF94025.1 KRF94027.1 GANO01001893 JAB57978.1 CH940660 KRF78087.1 GDAI01000983 JAI16620.1 EDW07387.1 GGFK01003449 MBW36770.1 AJWK01018121 AJWK01018122 GGFJ01002495 MBW51636.1 EDW58047.2 AE014298 AY061021 AY118309 BT099930 BT099932 BT126034 CH963925 KRF98851.1 CH954180 KQS29802.1 GEDC01018942 JAS18356.1 CM000162 KRK06485.1 DS235873 EEB19620.1 GFDF01003611 JAV10473.1 CH916370 EDV99533.1 EDV46049.2 GAPW01000244 JAC13354.1 CH478033 EAT34274.1 OUUW01000039 SPP89913.1 EDX01971.2 GFDL01014463 JAV20582.1 EDW78327.1
JAT83755.1 AGBW02012593 OWR44781.1 ODYU01007655 SOQ50622.1 PCG66897.1 KQ459594 KPI96110.1 KQ461108 KPJ09273.1 NEVH01002994 PNF40912.1 ADTU01011953 KQ976396 KYM92988.1 KQ981727 KYN36971.1 KK852685 KDR18482.1 GL440682 EFN65572.1 PNF40910.1 QOIP01000010 RLU18055.1 GEBQ01024317 GEBQ01003623 JAT15660.1 JAT36354.1 KQ435922 KOX68640.1 KZ288232 PBC31530.1 KQ982490 KYQ55788.1 KQ414632 KOC67123.1 NNAY01001051 OXU25331.1 GL449158 EFN83192.1 GEDC01030221 JAS07077.1 AAZX01000334 GEBQ01026154 GEBQ01024264 JAT13823.1 JAT15713.1 KK107167 EZA56367.1 KQ971327 EFA01267.1 GFTR01007979 JAW08447.1 GEDC01000906 JAS36392.1 ABLF02040575 ABLF02040576 GL764129 EFZ18387.1 GEZM01076826 JAV63615.1 APGK01043126 APGK01043127 APGK01043128 APGK01043129 KB741011 ENN75573.1 GBYB01011036 JAG80803.1 KB631957 ERL87468.1 GDHC01020521 JAP98107.1 CVRI01000059 CRL03768.1 AAAB01008859 EAA08217.5 GDHC01016241 JAQ02388.1 CRL03769.1 APCN01000435 EGK96918.1 GECL01001202 JAP04922.1 GGMR01005871 MBY18490.1 GDHC01010247 JAQ08382.1 GGFJ01002226 MBW51367.1 ADMH02001618 ETN61773.1 GGFM01001288 MBW22039.1 ATLV01013859 KE524905 KFB38212.1 AXCN02000052 GEBQ01031355 GEBQ01005695 JAT08622.1 JAT34282.1 GEDC01001382 JAS35916.1 GL888417 EGI61310.1 ACPB03020531 CH933810 KRF94026.1 GGFK01003230 MBW36551.1 KRF94025.1 KRF94027.1 GANO01001893 JAB57978.1 CH940660 KRF78087.1 GDAI01000983 JAI16620.1 EDW07387.1 GGFK01003449 MBW36770.1 AJWK01018121 AJWK01018122 GGFJ01002495 MBW51636.1 EDW58047.2 AE014298 AY061021 AY118309 BT099930 BT099932 BT126034 CH963925 KRF98851.1 CH954180 KQS29802.1 GEDC01018942 JAS18356.1 CM000162 KRK06485.1 DS235873 EEB19620.1 GFDF01003611 JAV10473.1 CH916370 EDV99533.1 EDV46049.2 GAPW01000244 JAC13354.1 CH478033 EAT34274.1 OUUW01000039 SPP89913.1 EDX01971.2 GFDL01014463 JAV20582.1 EDW78327.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000235965
+ More
UP000005205 UP000078540 UP000078541 UP000027135 UP000000311 UP000279307 UP000053105 UP000242457 UP000075809 UP000053825 UP000215335 UP000008237 UP000002358 UP000053097 UP000007266 UP000007819 UP000075903 UP000075902 UP000076407 UP000075882 UP000019118 UP000030742 UP000183832 UP000007062 UP000075840 UP000000673 UP000069272 UP000192223 UP000030765 UP000075900 UP000075886 UP000076408 UP000075881 UP000075880 UP000007755 UP000075884 UP000075920 UP000015103 UP000075885 UP000009192 UP000008792 UP000092461 UP000000803 UP000007798 UP000008711 UP000002282 UP000009046 UP000001070 UP000008820 UP000268350 UP000192221
UP000005205 UP000078540 UP000078541 UP000027135 UP000000311 UP000279307 UP000053105 UP000242457 UP000075809 UP000053825 UP000215335 UP000008237 UP000002358 UP000053097 UP000007266 UP000007819 UP000075903 UP000075902 UP000076407 UP000075882 UP000019118 UP000030742 UP000183832 UP000007062 UP000075840 UP000000673 UP000069272 UP000192223 UP000030765 UP000075900 UP000075886 UP000076408 UP000075881 UP000075880 UP000007755 UP000075884 UP000075920 UP000015103 UP000075885 UP000009192 UP000008792 UP000092461 UP000000803 UP000007798 UP000008711 UP000002282 UP000009046 UP000001070 UP000008820 UP000268350 UP000192221
Pfam
PF00566 RabGAP-TBC
SUPFAM
SSF47923
SSF47923
ProteinModelPortal
H9ITG0
A0A2A4J4U2
A0A2A4J634
A0A1E1W9Y6
A0A212ETK5
A0A2H1WCQ3
+ More
A0A2A4J422 A0A194PY85 A0A194QUR6 A0A2J7RJB0 A0A158NCK4 A0A195BYH3 A0A195F9B5 A0A067RH37 E2ALY0 A0A2J7RJA0 A0A3L8DC59 A0A1B6MKB5 A0A0M8ZPT1 A0A2A3EIJ2 A0A151X684 A0A0L7R8G4 A0A232F3K9 E2BM66 A0A1B6C0N9 K7J5B5 A0A1B6KW95 A0A026WJY9 D6WFG7 A0A224X7U8 A0A1B6EEN8 X1WJP3 E9IMA2 A0A1Y1KV77 A0A182UTN2 A0A182TJF7 A0A182XIC9 A0A182LFG3 N6TD12 A0A0C9RTX4 U4U569 A0A146KNV6 A0A1J1IUI5 Q7QBX8 A0A146L1Y0 A0A1J1IYV9 A0A182HTB4 F5HK77 A0A0V0GC26 A0A2S2NMS2 A0A146LNM6 A0A2M4BEC2 W5JEK1 A0A2M3Z0I4 A0A182F8N7 A0A1W4XH45 A0A1W4XSP3 A0A084VJR8 A0A182RCW4 A0A182Q3D9 A0A1B6MEE3 A0A182Y798 A0A182K833 A0A1B6EDA2 A0A182INU1 F4WX67 A0A182N4N1 A0A182VQ95 T1HVV6 A0A1W4XSK6 A0A182PA27 A0A1W4XRP5 A0A0Q9WZU9 A0A2M4A727 A0A0Q9WP52 U5EYP4 A0A0Q9W711 A0A0K8TRU0 B4L4A9 A0A2M4A7R0 A0A1B0CM39 A0A2M4BEZ8 B4MCX5 Q9VYY9-4 A0A0Q9WRJ7 A0A0Q5TIT0 A0A1B6CY08 A0A0R1EBB0 E0W1N8 Q9VYY9 A0A1L8DVN3 B4JIW9 B3NUD4 A0A023EXU8 Q16J34 A0A3B0K982 B4PYA9 A0A1W4UNL1 A0A1Q3EZ55 B4N1T8
A0A2A4J422 A0A194PY85 A0A194QUR6 A0A2J7RJB0 A0A158NCK4 A0A195BYH3 A0A195F9B5 A0A067RH37 E2ALY0 A0A2J7RJA0 A0A3L8DC59 A0A1B6MKB5 A0A0M8ZPT1 A0A2A3EIJ2 A0A151X684 A0A0L7R8G4 A0A232F3K9 E2BM66 A0A1B6C0N9 K7J5B5 A0A1B6KW95 A0A026WJY9 D6WFG7 A0A224X7U8 A0A1B6EEN8 X1WJP3 E9IMA2 A0A1Y1KV77 A0A182UTN2 A0A182TJF7 A0A182XIC9 A0A182LFG3 N6TD12 A0A0C9RTX4 U4U569 A0A146KNV6 A0A1J1IUI5 Q7QBX8 A0A146L1Y0 A0A1J1IYV9 A0A182HTB4 F5HK77 A0A0V0GC26 A0A2S2NMS2 A0A146LNM6 A0A2M4BEC2 W5JEK1 A0A2M3Z0I4 A0A182F8N7 A0A1W4XH45 A0A1W4XSP3 A0A084VJR8 A0A182RCW4 A0A182Q3D9 A0A1B6MEE3 A0A182Y798 A0A182K833 A0A1B6EDA2 A0A182INU1 F4WX67 A0A182N4N1 A0A182VQ95 T1HVV6 A0A1W4XSK6 A0A182PA27 A0A1W4XRP5 A0A0Q9WZU9 A0A2M4A727 A0A0Q9WP52 U5EYP4 A0A0Q9W711 A0A0K8TRU0 B4L4A9 A0A2M4A7R0 A0A1B0CM39 A0A2M4BEZ8 B4MCX5 Q9VYY9-4 A0A0Q9WRJ7 A0A0Q5TIT0 A0A1B6CY08 A0A0R1EBB0 E0W1N8 Q9VYY9 A0A1L8DVN3 B4JIW9 B3NUD4 A0A023EXU8 Q16J34 A0A3B0K982 B4PYA9 A0A1W4UNL1 A0A1Q3EZ55 B4N1T8
PDB
3HZJ
E-value=1.32037e-55,
Score=551
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Endosome
Endosome
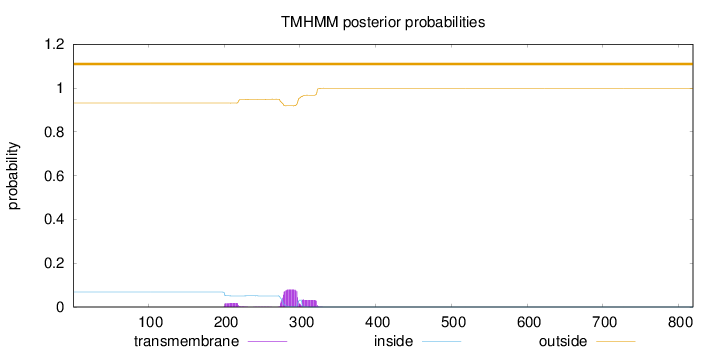
Length:
819
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.68957
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06810
outside
1 - 819
Population Genetic Test Statistics
Pi
19.120682
Theta
17.329432
Tajima's D
-1.234414
CLR
0.004277
CSRT
0.0978951052447378
Interpretation
Uncertain
