Pre Gene Modal
BGIBMGA000683
Annotation
PREDICTED:_beta-parvin_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 2.199 Nuclear Reliability : 1.511
Sequence
CDS
ATGTCTTCACCACGCCCAAAATCACCTCGTACTCCTGTGCTTCCCAAAAAAGAGGAAAAAGAGTCCTTTTGGGACAAAATTGGTACAATAGGGCGCAAAAAAGGGATTAAAGAAGTTCAAGATGTACAAGCTGAAGGGAAATATGCTATTGATTCTCCAGGCAGTCCAACTGCACCAGAGATTCCCCCTGAAGAATATACTTTGAATGAGAACGAAGAAAGGGCTATTATCGAACCTCGATCACTTGAAGATCCAAGGGTCAGAGATCTGATTCAAGTCTTGATTGACTGGATCAACGATGAACTAGCTTCACAGAGAATAATCGTCCAGGACATCATCGAAGATCTTTATGACGGTCAAGTGCTTCAAAAACTTTTAGAACAATTAACAAAGACGAAACTTGATGTACCTGAAGTAACTCAATCAGAAGAAGGACAGCGCCAGAAATTGACTATAGTTTTACGGGCTGTTAATCGAGTCTTGTACGGTACATCAAAACCAGTACCAAAATGGAGTGTGGATTCGATACATTCAAAAAGTATTGTTTCGATTCTTCATTTGCTCGTTGCTCTGGCCCGTCATTTCCGAGCTCCCATACGTCTCCCCGAAAATGTTTCTGTGAACGTTGTTATCGTCAAGAAAAACACACCGACCCAACTATCTCACCGGACTTTAATCGAAACCATAACGACTACTTATGACGATCTCGGCATGAAATGTGAACGTGATGCGTTTGATGCGTTGTTCGACCATGCACCGGACAAGCTCTTAGTTGTTAAGAAGTCACTTATTACATTTGTGAATAAACATTTGAGCAAAGTGAACTTAGAGGTAATGGATTTGGACACCCAGTTCCATGACGGAGTATACTTATGTCTTTTAATGGGCTTACTTGAAGAATTCTTTGTACCTCTTTACGACTTTCATTTAACACCTCAAGATTTTGATCAGAAAGTTCATAATGTATCATTTGCATTTGAGCTTATGCAAGATGTTGGTCTGGGAAAGCCCAAAGCCAGGCCTGAAGATATCGTGAATCTCGACCTCAAGTCAACCCTTCGAGTGCTTTACAATCTGTTCACAAAATATAAAAATATGGCTTAA
Protein
MSSPRPKSPRTPVLPKKEEKESFWDKIGTIGRKKGIKEVQDVQAEGKYAIDSPGSPTAPEIPPEEYTLNENEERAIIEPRSLEDPRVRDLIQVLIDWINDELASQRIIVQDIIEDLYDGQVLQKLLEQLTKTKLDVPEVTQSEEGQRQKLTIVLRAVNRVLYGTSKPVPKWSVDSIHSKSIVSILHLLVALARHFRAPIRLPENVSVNVVIVKKNTPTQLSHRTLIETITTTYDDLGMKCERDAFDALFDHAPDKLLVVKKSLITFVNKHLSKVNLEVMDLDTQFHDGVYLCLLMGLLEEFFVPLYDFHLTPQDFDQKVHNVSFAFELMQDVGLGKPKARPEDIVNLDLKSTLRVLYNLFTKYKNMA
Summary
Uniprot
H9ITV3
A0A2H1WC16
A0A194R0A4
A0A194PT19
A0A1E1WEJ5
A0A212ETG9
+ More
A0A0L7KJR2 A0A067QJH6 A0A1B6CD95 A0A2J7QJA1 D6WIR7 A0A1Y1L503 A0A1B6ER15 A0A146LGW1 A0A0A9XPL5 A0A1B6HKX8 A0A1B6JRL6 A0A026WN02 E2AFR6 A0A0J7L5P1 E2BWM8 A0A195B5T2 A0A151JNK5 A0A158NQA0 A0A1D2MXB1 F4WEU9 A0A1W4XHI8 A0A151X9T8 A0A151K1C8 A0A195CUL9 V5G4K4 A0A1L8E5K6 A0A224XEL8 A0A226E654 A0A0P5PFG5 A0A023F922 E9FRH7 R4G8R5 E0W1R1 A0A0P4W1D6 A0A0P6D0R0 A0A0P5ARH3 A0A0P6ETY5 A0A0P5K9P8 A0A0P6D6M1 A0A0P6BUW1 A0A0P5Q8D3 A0A0P4YP31 U5EVI0 K7J638 A0A232F1Z3 A0A0P5U859 A0A1L8E598 A0A0P6EFF3 A0A0P6DMX7 A0A0P5KXK7 A0A0C9R753 A0A0P4Z5U8 A0A0C9QGJ4 A0A0P5U3G9 C4WWS0 A0A0P4Z6Q5 A0A0P5Z7Y3 A0A1L8E587 A0A0N8DFY6 A0A0P6AC22 A0A2H8TKL0 A0A0N8C427 A0A0P6FLN8 A0A0P6I3B0 A0A0P5KES6 A0A0N8BA49 A0A0P5WAH7 A0A0P5L6T1 A0A0P5LLL5 A0A2S2R3I1 A0A0P5BIC0 A0A0N8A5P3 A0A0P5BQC9 A0A0P5V580 A0A0P5JFW2 A0A131Y326 A0A131XEG7 A0A0P6CNU6 A0A023FL22 A0A023GJ37 G3MM55 A0A182R1B1 A0A0P6FHM5 L7M933 A0A0N8CXM3 A0A224YSV7 A0A131Z2B7 A0A154NYR9 A0A0L7QSF4 A0A088A3I9 A0A0T6B079 A0A0M9A947 A0A2A3E3C7 A0A023FLY6
A0A0L7KJR2 A0A067QJH6 A0A1B6CD95 A0A2J7QJA1 D6WIR7 A0A1Y1L503 A0A1B6ER15 A0A146LGW1 A0A0A9XPL5 A0A1B6HKX8 A0A1B6JRL6 A0A026WN02 E2AFR6 A0A0J7L5P1 E2BWM8 A0A195B5T2 A0A151JNK5 A0A158NQA0 A0A1D2MXB1 F4WEU9 A0A1W4XHI8 A0A151X9T8 A0A151K1C8 A0A195CUL9 V5G4K4 A0A1L8E5K6 A0A224XEL8 A0A226E654 A0A0P5PFG5 A0A023F922 E9FRH7 R4G8R5 E0W1R1 A0A0P4W1D6 A0A0P6D0R0 A0A0P5ARH3 A0A0P6ETY5 A0A0P5K9P8 A0A0P6D6M1 A0A0P6BUW1 A0A0P5Q8D3 A0A0P4YP31 U5EVI0 K7J638 A0A232F1Z3 A0A0P5U859 A0A1L8E598 A0A0P6EFF3 A0A0P6DMX7 A0A0P5KXK7 A0A0C9R753 A0A0P4Z5U8 A0A0C9QGJ4 A0A0P5U3G9 C4WWS0 A0A0P4Z6Q5 A0A0P5Z7Y3 A0A1L8E587 A0A0N8DFY6 A0A0P6AC22 A0A2H8TKL0 A0A0N8C427 A0A0P6FLN8 A0A0P6I3B0 A0A0P5KES6 A0A0N8BA49 A0A0P5WAH7 A0A0P5L6T1 A0A0P5LLL5 A0A2S2R3I1 A0A0P5BIC0 A0A0N8A5P3 A0A0P5BQC9 A0A0P5V580 A0A0P5JFW2 A0A131Y326 A0A131XEG7 A0A0P6CNU6 A0A023FL22 A0A023GJ37 G3MM55 A0A182R1B1 A0A0P6FHM5 L7M933 A0A0N8CXM3 A0A224YSV7 A0A131Z2B7 A0A154NYR9 A0A0L7QSF4 A0A088A3I9 A0A0T6B079 A0A0M9A947 A0A2A3E3C7 A0A023FLY6
Pubmed
EMBL
BABH01007029
ODYU01007654
SOQ50621.1
KQ461108
KPJ09271.1
KQ459594
+ More
KPI96113.1 GDQN01011594 GDQN01005651 JAT79460.1 JAT85403.1 AGBW02012593 OWR44780.1 JTDY01009393 KOB63572.1 KK853417 KDR07747.1 GEDC01025869 JAS11429.1 NEVH01013559 PNF28639.1 KQ971321 EEZ99501.1 GEZM01064492 JAV68762.1 GECZ01029367 GECZ01026830 GECZ01001898 JAS40402.1 JAS42939.1 JAS67871.1 GDHC01012024 JAQ06605.1 GBHO01020952 JAG22652.1 GECU01032440 GECU01032370 GECU01022343 GECU01011483 JAS75266.1 JAS75336.1 JAS85363.1 JAS96223.1 GECU01005847 JAT01860.1 KK107167 QOIP01000010 EZA56484.1 RLU18139.1 GL439124 EFN67727.1 LBMM01000672 KMQ97814.1 GL451165 EFN79883.1 KQ976595 KYM79630.1 KQ978820 KYN28137.1 ADTU01022993 LJIJ01000420 ODM97676.1 GL888111 EGI67266.1 KQ982365 KYQ57132.1 KQ981189 KYN45228.1 KQ977279 KYN04376.1 GALX01003526 JAB64940.1 GFDF01000175 JAV13909.1 GFTR01005511 JAW10915.1 LNIX01000006 OXA53093.1 GDIQ01135344 JAL16382.1 GBBI01000746 JAC17966.1 GL732523 EFX90168.1 ACPB03012309 GAHY01000512 JAA76998.1 DS235873 EEB19643.1 GDKW01001112 JAI55483.1 GDIQ01084615 JAN10122.1 GDIP01195090 JAJ28312.1 GDIQ01059113 JAN35624.1 GDIQ01192719 JAK59006.1 GDIQ01089895 JAN04842.1 GDIP01024713 JAM79002.1 GDIQ01120838 JAL30888.1 GDIP01224834 JAI98567.1 GANO01001871 JAB58000.1 NNAY01001275 OXU24530.1 GDIP01120494 JAL83220.1 GFDF01000206 JAV13878.1 GDIQ01064571 JAN30166.1 GDIQ01089896 JAN04841.1 GDIQ01178618 JAK73107.1 GBYB01002591 JAG72358.1 GDIP01219093 JAJ04309.1 GBYB01002587 GBYB01002589 JAG72354.1 JAG72356.1 GDIP01118403 JAL85311.1 ABLF02035889 AK342232 BAH72340.1 GDIP01221490 JAJ01912.1 GDIP01048765 JAM54950.1 GFDF01000174 JAV13910.1 GDIP01037559 JAM66156.1 GDIP01036460 JAM67255.1 GFXV01002013 MBW13818.1 GDIQ01116843 JAL34883.1 GDIQ01047681 JAN47056.1 GDIQ01009967 JAN84770.1 GDIQ01210960 JAK40765.1 GDIQ01197867 JAK53858.1 GDIP01088864 JAM14851.1 GDIQ01175072 JAK76653.1 GDIQ01192720 JAK59005.1 GGMS01015373 MBY84576.1 GDIP01184687 JAJ38715.1 GDIP01179993 JAJ43409.1 GDIP01181798 JAJ41604.1 GDIP01104702 JAL99012.1 GDIQ01225305 JAK26420.1 GEFM01002888 JAP72908.1 GEFH01004540 JAP64041.1 GDIQ01089897 JAN04840.1 GBBK01002772 JAC21710.1 GBBM01001476 JAC33942.1 JO842956 AEO34573.1 AXCN02000052 GDIQ01047680 JAN47057.1 GACK01004647 JAA60387.1 GDIP01088863 JAM14852.1 GFPF01005928 MAA17074.1 GEDV01003130 JAP85427.1 KQ434783 KZC04712.1 KQ414758 KOC61484.1 LJIG01016378 KRT80780.1 KQ435724 KOX78473.1 KZ288403 PBC26227.1 GBBK01002789 JAC21693.1
KPI96113.1 GDQN01011594 GDQN01005651 JAT79460.1 JAT85403.1 AGBW02012593 OWR44780.1 JTDY01009393 KOB63572.1 KK853417 KDR07747.1 GEDC01025869 JAS11429.1 NEVH01013559 PNF28639.1 KQ971321 EEZ99501.1 GEZM01064492 JAV68762.1 GECZ01029367 GECZ01026830 GECZ01001898 JAS40402.1 JAS42939.1 JAS67871.1 GDHC01012024 JAQ06605.1 GBHO01020952 JAG22652.1 GECU01032440 GECU01032370 GECU01022343 GECU01011483 JAS75266.1 JAS75336.1 JAS85363.1 JAS96223.1 GECU01005847 JAT01860.1 KK107167 QOIP01000010 EZA56484.1 RLU18139.1 GL439124 EFN67727.1 LBMM01000672 KMQ97814.1 GL451165 EFN79883.1 KQ976595 KYM79630.1 KQ978820 KYN28137.1 ADTU01022993 LJIJ01000420 ODM97676.1 GL888111 EGI67266.1 KQ982365 KYQ57132.1 KQ981189 KYN45228.1 KQ977279 KYN04376.1 GALX01003526 JAB64940.1 GFDF01000175 JAV13909.1 GFTR01005511 JAW10915.1 LNIX01000006 OXA53093.1 GDIQ01135344 JAL16382.1 GBBI01000746 JAC17966.1 GL732523 EFX90168.1 ACPB03012309 GAHY01000512 JAA76998.1 DS235873 EEB19643.1 GDKW01001112 JAI55483.1 GDIQ01084615 JAN10122.1 GDIP01195090 JAJ28312.1 GDIQ01059113 JAN35624.1 GDIQ01192719 JAK59006.1 GDIQ01089895 JAN04842.1 GDIP01024713 JAM79002.1 GDIQ01120838 JAL30888.1 GDIP01224834 JAI98567.1 GANO01001871 JAB58000.1 NNAY01001275 OXU24530.1 GDIP01120494 JAL83220.1 GFDF01000206 JAV13878.1 GDIQ01064571 JAN30166.1 GDIQ01089896 JAN04841.1 GDIQ01178618 JAK73107.1 GBYB01002591 JAG72358.1 GDIP01219093 JAJ04309.1 GBYB01002587 GBYB01002589 JAG72354.1 JAG72356.1 GDIP01118403 JAL85311.1 ABLF02035889 AK342232 BAH72340.1 GDIP01221490 JAJ01912.1 GDIP01048765 JAM54950.1 GFDF01000174 JAV13910.1 GDIP01037559 JAM66156.1 GDIP01036460 JAM67255.1 GFXV01002013 MBW13818.1 GDIQ01116843 JAL34883.1 GDIQ01047681 JAN47056.1 GDIQ01009967 JAN84770.1 GDIQ01210960 JAK40765.1 GDIQ01197867 JAK53858.1 GDIP01088864 JAM14851.1 GDIQ01175072 JAK76653.1 GDIQ01192720 JAK59005.1 GGMS01015373 MBY84576.1 GDIP01184687 JAJ38715.1 GDIP01179993 JAJ43409.1 GDIP01181798 JAJ41604.1 GDIP01104702 JAL99012.1 GDIQ01225305 JAK26420.1 GEFM01002888 JAP72908.1 GEFH01004540 JAP64041.1 GDIQ01089897 JAN04840.1 GBBK01002772 JAC21710.1 GBBM01001476 JAC33942.1 JO842956 AEO34573.1 AXCN02000052 GDIQ01047680 JAN47057.1 GACK01004647 JAA60387.1 GDIP01088863 JAM14852.1 GFPF01005928 MAA17074.1 GEDV01003130 JAP85427.1 KQ434783 KZC04712.1 KQ414758 KOC61484.1 LJIG01016378 KRT80780.1 KQ435724 KOX78473.1 KZ288403 PBC26227.1 GBBK01002789 JAC21693.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000027135
+ More
UP000235965 UP000007266 UP000053097 UP000279307 UP000000311 UP000036403 UP000008237 UP000078540 UP000078492 UP000005205 UP000094527 UP000007755 UP000192223 UP000075809 UP000078541 UP000078542 UP000198287 UP000000305 UP000015103 UP000009046 UP000002358 UP000215335 UP000007819 UP000075886 UP000076502 UP000053825 UP000005203 UP000053105 UP000242457
UP000235965 UP000007266 UP000053097 UP000279307 UP000000311 UP000036403 UP000008237 UP000078540 UP000078492 UP000005205 UP000094527 UP000007755 UP000192223 UP000075809 UP000078541 UP000078542 UP000198287 UP000000305 UP000015103 UP000009046 UP000002358 UP000215335 UP000007819 UP000075886 UP000076502 UP000053825 UP000005203 UP000053105 UP000242457
PRIDE
Pfam
PF00307 CH
SUPFAM
SSF47576
SSF47576
Gene 3D
ProteinModelPortal
H9ITV3
A0A2H1WC16
A0A194R0A4
A0A194PT19
A0A1E1WEJ5
A0A212ETG9
+ More
A0A0L7KJR2 A0A067QJH6 A0A1B6CD95 A0A2J7QJA1 D6WIR7 A0A1Y1L503 A0A1B6ER15 A0A146LGW1 A0A0A9XPL5 A0A1B6HKX8 A0A1B6JRL6 A0A026WN02 E2AFR6 A0A0J7L5P1 E2BWM8 A0A195B5T2 A0A151JNK5 A0A158NQA0 A0A1D2MXB1 F4WEU9 A0A1W4XHI8 A0A151X9T8 A0A151K1C8 A0A195CUL9 V5G4K4 A0A1L8E5K6 A0A224XEL8 A0A226E654 A0A0P5PFG5 A0A023F922 E9FRH7 R4G8R5 E0W1R1 A0A0P4W1D6 A0A0P6D0R0 A0A0P5ARH3 A0A0P6ETY5 A0A0P5K9P8 A0A0P6D6M1 A0A0P6BUW1 A0A0P5Q8D3 A0A0P4YP31 U5EVI0 K7J638 A0A232F1Z3 A0A0P5U859 A0A1L8E598 A0A0P6EFF3 A0A0P6DMX7 A0A0P5KXK7 A0A0C9R753 A0A0P4Z5U8 A0A0C9QGJ4 A0A0P5U3G9 C4WWS0 A0A0P4Z6Q5 A0A0P5Z7Y3 A0A1L8E587 A0A0N8DFY6 A0A0P6AC22 A0A2H8TKL0 A0A0N8C427 A0A0P6FLN8 A0A0P6I3B0 A0A0P5KES6 A0A0N8BA49 A0A0P5WAH7 A0A0P5L6T1 A0A0P5LLL5 A0A2S2R3I1 A0A0P5BIC0 A0A0N8A5P3 A0A0P5BQC9 A0A0P5V580 A0A0P5JFW2 A0A131Y326 A0A131XEG7 A0A0P6CNU6 A0A023FL22 A0A023GJ37 G3MM55 A0A182R1B1 A0A0P6FHM5 L7M933 A0A0N8CXM3 A0A224YSV7 A0A131Z2B7 A0A154NYR9 A0A0L7QSF4 A0A088A3I9 A0A0T6B079 A0A0M9A947 A0A2A3E3C7 A0A023FLY6
A0A0L7KJR2 A0A067QJH6 A0A1B6CD95 A0A2J7QJA1 D6WIR7 A0A1Y1L503 A0A1B6ER15 A0A146LGW1 A0A0A9XPL5 A0A1B6HKX8 A0A1B6JRL6 A0A026WN02 E2AFR6 A0A0J7L5P1 E2BWM8 A0A195B5T2 A0A151JNK5 A0A158NQA0 A0A1D2MXB1 F4WEU9 A0A1W4XHI8 A0A151X9T8 A0A151K1C8 A0A195CUL9 V5G4K4 A0A1L8E5K6 A0A224XEL8 A0A226E654 A0A0P5PFG5 A0A023F922 E9FRH7 R4G8R5 E0W1R1 A0A0P4W1D6 A0A0P6D0R0 A0A0P5ARH3 A0A0P6ETY5 A0A0P5K9P8 A0A0P6D6M1 A0A0P6BUW1 A0A0P5Q8D3 A0A0P4YP31 U5EVI0 K7J638 A0A232F1Z3 A0A0P5U859 A0A1L8E598 A0A0P6EFF3 A0A0P6DMX7 A0A0P5KXK7 A0A0C9R753 A0A0P4Z5U8 A0A0C9QGJ4 A0A0P5U3G9 C4WWS0 A0A0P4Z6Q5 A0A0P5Z7Y3 A0A1L8E587 A0A0N8DFY6 A0A0P6AC22 A0A2H8TKL0 A0A0N8C427 A0A0P6FLN8 A0A0P6I3B0 A0A0P5KES6 A0A0N8BA49 A0A0P5WAH7 A0A0P5L6T1 A0A0P5LLL5 A0A2S2R3I1 A0A0P5BIC0 A0A0N8A5P3 A0A0P5BQC9 A0A0P5V580 A0A0P5JFW2 A0A131Y326 A0A131XEG7 A0A0P6CNU6 A0A023FL22 A0A023GJ37 G3MM55 A0A182R1B1 A0A0P6FHM5 L7M933 A0A0N8CXM3 A0A224YSV7 A0A131Z2B7 A0A154NYR9 A0A0L7QSF4 A0A088A3I9 A0A0T6B079 A0A0M9A947 A0A2A3E3C7 A0A023FLY6
PDB
4EDN
E-value=9.99527e-55,
Score=539
Ontologies
GO
PANTHER
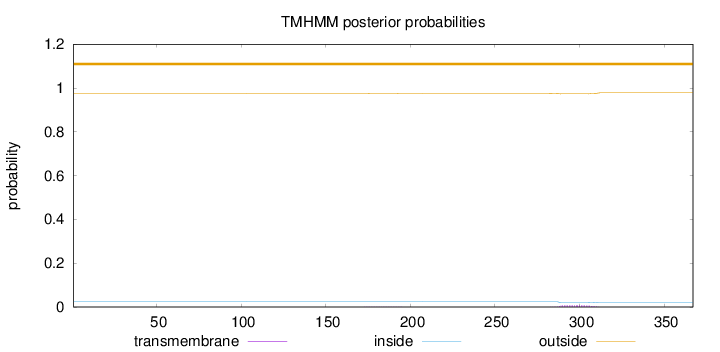
Topology
Length:
367
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15816
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02422
outside
1 - 367
Population Genetic Test Statistics
Pi
6.198229
Theta
17.258957
Tajima's D
-1.56155
CLR
0.36793
CSRT
0.0554472276386181
Interpretation
Uncertain
