Gene
KWMTBOMO00301
Pre Gene Modal
BGIBMGA000541
Annotation
PREDICTED:_lipopolysaccharide-induced_tumor_necrosis_factor-alpha_factor_homolog_[Plutella_xylostella]
Transcription factor
Location in the cell
Extracellular Reliability : 3.549
Sequence
CDS
ATGGAAAAAAATGGCAATCCTCCTCCATATGTATGGGGCCAGGCAAATGTTTTACAGCCTCCGCCTGCTGCTCCACCTAGCTACTCGCAAGCAGTGGGGGGTGTAGGGCCTTCCAGCCCTTACACACCTCAGTACCCAACAACAACAGGGCCACAAATTGTTACAACCGTAGTACCGGTTGGGCCGCATCCAACTCATATGATTTGTCCTAGTTGCCATGCCGAAATTAACACATCAACACAAACAAAACCTGGTCTCATTGCATACATAACTGGTGCAATTATATTCCTTTTAGGATGTATATGTGGGTGCTGCTTGATTCCCTGCTGTATTGACAGCTGTATGGACGTACACCATAAATGTCCTCATTGCGATACTTACCTCGGTCGCTATCGTCGATAA
Protein
MEKNGNPPPYVWGQANVLQPPPAAPPSYSQAVGGVGPSSPYTPQYPTTTGPQIVTTVVPVGPHPTHMICPSCHAEINTSTQTKPGLIAYITGAIIFLLGCICGCCLIPCCIDSCMDVHHKCPHCDTYLGRYRR
Summary
Uniprot
H9ITG1
A0A212ETH5
A0A2H1WC33
A0A194QWD0
I4DP79
I4DR29
+ More
T1E7L9 A0A2M4CTG3 A0A2M3Z8R2 A0A2M4AFW9 A0A2M4C3V4 A0A182FEP3 A0A182HQI4 Q7PI09 A0A182WYY3 A0A182K4A7 A0A182N212 U5ESB5 B0WT89 A0A1Q3FCM7 A0A084WCX0 A0A182IQ94 A0A182YKQ3 A0A067RLV9 B4N6B9 A0A087ZUA4 A0A1A9VMW7 A0A1A9Z8X8 D3TLV8 A0A1A9WCY7 A0A2A3ELC4 V9IKT6 A0A0M8ZUP1 A0A182WLZ0 B3NF39 B4PCU3 Q16RP8 A0A1I8NKD4 B3M7D2 B4LFE8 A0A023EFK3 A0A182PEN8 A0A069DVW9 A0A3B0J437 Q29DK8 B4H6V0 A0A0Q9WJT0 B4KZZ1 A0A2J7RKB4 A0A0A1X3G8 A0A034WKC2 A0A0K8VA26 B4J346 A0A0V0G912 A0A182MMT2 E2BEJ5 K7J9N5 A0A026W933 E9J3H4 A0A0J7KUN1 A0A151JN35 F4WJ13 A0A195FPY1 A0A151WX34 D1FQ30 A0A1S3DI77 A0A195CTM9 A0A195BMJ0 A0A158NTR8 A0A0A9ZGQ8 A0A224XWG8 A0A1J1IEB1
T1E7L9 A0A2M4CTG3 A0A2M3Z8R2 A0A2M4AFW9 A0A2M4C3V4 A0A182FEP3 A0A182HQI4 Q7PI09 A0A182WYY3 A0A182K4A7 A0A182N212 U5ESB5 B0WT89 A0A1Q3FCM7 A0A084WCX0 A0A182IQ94 A0A182YKQ3 A0A067RLV9 B4N6B9 A0A087ZUA4 A0A1A9VMW7 A0A1A9Z8X8 D3TLV8 A0A1A9WCY7 A0A2A3ELC4 V9IKT6 A0A0M8ZUP1 A0A182WLZ0 B3NF39 B4PCU3 Q16RP8 A0A1I8NKD4 B3M7D2 B4LFE8 A0A023EFK3 A0A182PEN8 A0A069DVW9 A0A3B0J437 Q29DK8 B4H6V0 A0A0Q9WJT0 B4KZZ1 A0A2J7RKB4 A0A0A1X3G8 A0A034WKC2 A0A0K8VA26 B4J346 A0A0V0G912 A0A182MMT2 E2BEJ5 K7J9N5 A0A026W933 E9J3H4 A0A0J7KUN1 A0A151JN35 F4WJ13 A0A195FPY1 A0A151WX34 D1FQ30 A0A1S3DI77 A0A195CTM9 A0A195BMJ0 A0A158NTR8 A0A0A9ZGQ8 A0A224XWG8 A0A1J1IEB1
Pubmed
EMBL
BABH01007028
AGBW02012593
OWR44779.1
ODYU01007654
SOQ50620.1
KQ461108
+ More
KPJ09270.1 AK403460 KQ459594 BAM19719.1 KPI96114.1 AK404912 BAM20369.1 GAMD01002917 JAA98673.1 GGFL01004435 MBW68613.1 GGFM01004077 MBW24828.1 GGFK01006360 MBW39681.1 GGFJ01010700 MBW59841.1 APCN01004623 AAAB01008948 EAA44328.1 GANO01002433 JAB57438.1 DS232081 EDS34238.1 GFDL01009704 JAV25341.1 ATLV01022896 KE525337 KFB48064.1 KK852598 KDR20553.1 CH964154 EDW79908.1 CCAG010016976 EZ422410 ADD18686.1 KZ288223 PBC31959.1 JR050476 AEY61292.1 KQ435835 KOX71510.1 CH954178 EDV50381.1 KQS43247.1 CM000159 EDW93847.1 KRK01440.1 CH477700 EAT37088.1 CH902618 EDV39830.1 CH940647 EDW69246.1 KRF84261.1 JXUM01009249 JXUM01009250 JXUM01009251 JXUM01047777 JXUM01047778 JXUM01047779 JXUM01047780 GAPW01005992 JAC07606.1 GBGD01003445 JAC85444.1 OUUW01000002 SPP76524.1 CH379070 EAL30406.2 CH479215 EDW33538.1 KRF84262.1 CH933809 EDW18003.1 KRG05894.1 NEVH01002981 PNF41277.1 GBXI01008438 JAD05854.1 GAKP01004155 JAC54797.1 GDHF01033567 GDHF01028588 GDHF01023782 GDHF01016616 GDHF01009974 GDHF01008760 JAI18747.1 JAI23726.1 JAI28532.1 JAI35698.1 JAI42340.1 JAI43554.1 CH916366 EDV96117.1 GECL01001856 JAP04268.1 AXCM01005049 GL447817 EFN85885.1 AAZX01002711 KK107348 QOIP01000010 EZA52141.1 RLU17845.1 GL768066 EFZ12573.1 LBMM01003062 KMQ94011.1 KQ978878 KYN27860.1 GL888181 EGI65783.1 KQ981382 KYN42347.1 KQ982673 KYQ52440.1 EZ419930 ACZ28285.1 KQ977279 KYN04053.1 KQ976435 KYM87095.1 ADTU01026117 GBHO01000263 GDHC01021151 JAG43341.1 JAP97477.1 GFTR01002218 JAW14208.1 CVRI01000047 CRK98106.1
KPJ09270.1 AK403460 KQ459594 BAM19719.1 KPI96114.1 AK404912 BAM20369.1 GAMD01002917 JAA98673.1 GGFL01004435 MBW68613.1 GGFM01004077 MBW24828.1 GGFK01006360 MBW39681.1 GGFJ01010700 MBW59841.1 APCN01004623 AAAB01008948 EAA44328.1 GANO01002433 JAB57438.1 DS232081 EDS34238.1 GFDL01009704 JAV25341.1 ATLV01022896 KE525337 KFB48064.1 KK852598 KDR20553.1 CH964154 EDW79908.1 CCAG010016976 EZ422410 ADD18686.1 KZ288223 PBC31959.1 JR050476 AEY61292.1 KQ435835 KOX71510.1 CH954178 EDV50381.1 KQS43247.1 CM000159 EDW93847.1 KRK01440.1 CH477700 EAT37088.1 CH902618 EDV39830.1 CH940647 EDW69246.1 KRF84261.1 JXUM01009249 JXUM01009250 JXUM01009251 JXUM01047777 JXUM01047778 JXUM01047779 JXUM01047780 GAPW01005992 JAC07606.1 GBGD01003445 JAC85444.1 OUUW01000002 SPP76524.1 CH379070 EAL30406.2 CH479215 EDW33538.1 KRF84262.1 CH933809 EDW18003.1 KRG05894.1 NEVH01002981 PNF41277.1 GBXI01008438 JAD05854.1 GAKP01004155 JAC54797.1 GDHF01033567 GDHF01028588 GDHF01023782 GDHF01016616 GDHF01009974 GDHF01008760 JAI18747.1 JAI23726.1 JAI28532.1 JAI35698.1 JAI42340.1 JAI43554.1 CH916366 EDV96117.1 GECL01001856 JAP04268.1 AXCM01005049 GL447817 EFN85885.1 AAZX01002711 KK107348 QOIP01000010 EZA52141.1 RLU17845.1 GL768066 EFZ12573.1 LBMM01003062 KMQ94011.1 KQ978878 KYN27860.1 GL888181 EGI65783.1 KQ981382 KYN42347.1 KQ982673 KYQ52440.1 EZ419930 ACZ28285.1 KQ977279 KYN04053.1 KQ976435 KYM87095.1 ADTU01026117 GBHO01000263 GDHC01021151 JAG43341.1 JAP97477.1 GFTR01002218 JAW14208.1 CVRI01000047 CRK98106.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000069272
UP000075840
+ More
UP000007062 UP000076407 UP000075881 UP000075884 UP000002320 UP000030765 UP000075880 UP000076408 UP000027135 UP000007798 UP000005203 UP000078200 UP000092445 UP000092444 UP000091820 UP000242457 UP000053105 UP000075920 UP000008711 UP000002282 UP000008820 UP000095301 UP000007801 UP000008792 UP000069940 UP000075885 UP000268350 UP000001819 UP000008744 UP000009192 UP000235965 UP000001070 UP000075883 UP000008237 UP000002358 UP000053097 UP000279307 UP000036403 UP000078492 UP000007755 UP000078541 UP000075809 UP000079169 UP000078542 UP000078540 UP000005205 UP000183832
UP000007062 UP000076407 UP000075881 UP000075884 UP000002320 UP000030765 UP000075880 UP000076408 UP000027135 UP000007798 UP000005203 UP000078200 UP000092445 UP000092444 UP000091820 UP000242457 UP000053105 UP000075920 UP000008711 UP000002282 UP000008820 UP000095301 UP000007801 UP000008792 UP000069940 UP000075885 UP000268350 UP000001819 UP000008744 UP000009192 UP000235965 UP000001070 UP000075883 UP000008237 UP000002358 UP000053097 UP000279307 UP000036403 UP000078492 UP000007755 UP000078541 UP000075809 UP000079169 UP000078542 UP000078540 UP000005205 UP000183832
PRIDE
Pfam
PF10601 zf-LITAF-like
ProteinModelPortal
H9ITG1
A0A212ETH5
A0A2H1WC33
A0A194QWD0
I4DP79
I4DR29
+ More
T1E7L9 A0A2M4CTG3 A0A2M3Z8R2 A0A2M4AFW9 A0A2M4C3V4 A0A182FEP3 A0A182HQI4 Q7PI09 A0A182WYY3 A0A182K4A7 A0A182N212 U5ESB5 B0WT89 A0A1Q3FCM7 A0A084WCX0 A0A182IQ94 A0A182YKQ3 A0A067RLV9 B4N6B9 A0A087ZUA4 A0A1A9VMW7 A0A1A9Z8X8 D3TLV8 A0A1A9WCY7 A0A2A3ELC4 V9IKT6 A0A0M8ZUP1 A0A182WLZ0 B3NF39 B4PCU3 Q16RP8 A0A1I8NKD4 B3M7D2 B4LFE8 A0A023EFK3 A0A182PEN8 A0A069DVW9 A0A3B0J437 Q29DK8 B4H6V0 A0A0Q9WJT0 B4KZZ1 A0A2J7RKB4 A0A0A1X3G8 A0A034WKC2 A0A0K8VA26 B4J346 A0A0V0G912 A0A182MMT2 E2BEJ5 K7J9N5 A0A026W933 E9J3H4 A0A0J7KUN1 A0A151JN35 F4WJ13 A0A195FPY1 A0A151WX34 D1FQ30 A0A1S3DI77 A0A195CTM9 A0A195BMJ0 A0A158NTR8 A0A0A9ZGQ8 A0A224XWG8 A0A1J1IEB1
T1E7L9 A0A2M4CTG3 A0A2M3Z8R2 A0A2M4AFW9 A0A2M4C3V4 A0A182FEP3 A0A182HQI4 Q7PI09 A0A182WYY3 A0A182K4A7 A0A182N212 U5ESB5 B0WT89 A0A1Q3FCM7 A0A084WCX0 A0A182IQ94 A0A182YKQ3 A0A067RLV9 B4N6B9 A0A087ZUA4 A0A1A9VMW7 A0A1A9Z8X8 D3TLV8 A0A1A9WCY7 A0A2A3ELC4 V9IKT6 A0A0M8ZUP1 A0A182WLZ0 B3NF39 B4PCU3 Q16RP8 A0A1I8NKD4 B3M7D2 B4LFE8 A0A023EFK3 A0A182PEN8 A0A069DVW9 A0A3B0J437 Q29DK8 B4H6V0 A0A0Q9WJT0 B4KZZ1 A0A2J7RKB4 A0A0A1X3G8 A0A034WKC2 A0A0K8VA26 B4J346 A0A0V0G912 A0A182MMT2 E2BEJ5 K7J9N5 A0A026W933 E9J3H4 A0A0J7KUN1 A0A151JN35 F4WJ13 A0A195FPY1 A0A151WX34 D1FQ30 A0A1S3DI77 A0A195CTM9 A0A195BMJ0 A0A158NTR8 A0A0A9ZGQ8 A0A224XWG8 A0A1J1IEB1
Ontologies
GO
PANTHER
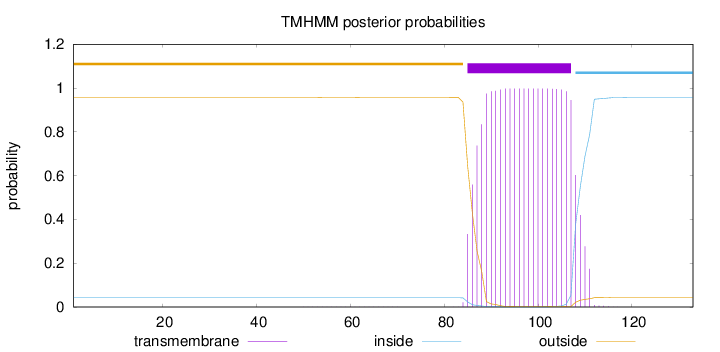
Topology
Length:
133
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.82184
Exp number, first 60 AAs:
0.00336
Total prob of N-in:
0.04230
outside
1 - 84
TMhelix
85 - 107
inside
108 - 133
Population Genetic Test Statistics
Pi
1.413096
Theta
5.004939
Tajima's D
-2.109181
CLR
1.472201
CSRT
0.00649967501624919
Interpretation
Uncertain
