Gene
KWMTBOMO00292
Pre Gene Modal
BGIBMGA011396
Annotation
endonuclease-reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.111
Sequence
CDS
ATGTTGATAGAGAAAAGGAAAAGACTAAAAGAACAAGGCATTGTAAACAATAGTCAAAGGGAGCGCTACAATCAATTGCATGCAGAGATTCAAAGGTCTACTCGGAGAGACAGAGATGAGCACATCAATAATATGTGTGAAGAAATTGAGGGACATGCTGATAAAGTGCAGACTAAGGACCTTTTCCTGAAAGTGAAGCAGCTAACGGGTGTCTTCAAGCCGAAGAAATGGATTATCGAAGATAGTAGAGGTAACACCTTGACAGAAATGGAACATGTTTTGGAGAGGTGGAGAAACTACTGTTATGACCTATATCGCGATGAAGATAATTTATGCACCGCCAAAATATGGGATGACCACCTGGAAGAGCCTCCTATACTACTTTCTGAGGTTGAATATGCAATACAAAAACTTAAGCATAAGGTGTTCATTGTCAAGCACAAAGAAAATTTAGATCTAAAATATTTCGATCTGGAGCACTCGCCTTCCAGTCACGAGTACCATAACTTGTCTTCGTTTTCCAAGACGCTGTACGAACAGAGCGGTTAA
Protein
MLIEKRKRLKEQGIVNNSQRERYNQLHAEIQRSTRRDRDEHINNMCEEIEGHADKVQTKDLFLKVKQLTGVFKPKKWIIEDSRGNTLTEMEHVLERWRNYCYDLYRDEDNLCTAKIWDDHLEEPPILLSEVEYAIQKLKHKVFIVKHKENLDLKYFDLEHSPSSHEYHNLSSFSKTLYEQSG
Summary
Uniprot
D5LB39
H9JCQ5
D7F163
A0A2W1BNH4
A0A0J7KGF7
A0A2W1BGD9
+ More
D7F174 A0A2H1W4B8 A0A3D1L484 D7F172 D7F168 D7F167 D7F162 A0A0L7KZZ5 D7F175 A0A0J7L4A4 A0A2G8JL44 A0A0L7LF38 A0A0J7K8A5 A0A0B1PP52 A0A085NA39 W5P980 A0A023G4L3 A0A023GD30 A0A2H6KKM9 H3A4I6 A0A1S4EQZ4 A0A2J7PHB6 A0A2J7PL22 H3J6U9 A0A2J7PPK7 A0A2J7PQR6 H3A3N0 A0A2J7RS89 A0A2J7PNJ4 J9KN99 A0A2S2QXQ0 A0A2J7RE76 A0A2J7R7C2 A0A2J7RGX9 A0A2J7QFW5 A0A2J7Q4I0 A0A2J7Q6D5 A0A2J7QDX3 A0A2J7RJY3 A0A2J7PTB8 A0A2J7R2Y0 A0A2J7PT05 A0A2J7QRB7 A0A2T7NW75 A0A2J7PSI0 A0A2J7Q6T5 A0A2J7PSH5 A0A2J7QEY3
D7F174 A0A2H1W4B8 A0A3D1L484 D7F172 D7F168 D7F167 D7F162 A0A0L7KZZ5 D7F175 A0A0J7L4A4 A0A2G8JL44 A0A0L7LF38 A0A0J7K8A5 A0A0B1PP52 A0A085NA39 W5P980 A0A023G4L3 A0A023GD30 A0A2H6KKM9 H3A4I6 A0A1S4EQZ4 A0A2J7PHB6 A0A2J7PL22 H3J6U9 A0A2J7PPK7 A0A2J7PQR6 H3A3N0 A0A2J7RS89 A0A2J7PNJ4 J9KN99 A0A2S2QXQ0 A0A2J7RE76 A0A2J7R7C2 A0A2J7RGX9 A0A2J7QFW5 A0A2J7Q4I0 A0A2J7Q6D5 A0A2J7QDX3 A0A2J7RJY3 A0A2J7PTB8 A0A2J7R2Y0 A0A2J7PT05 A0A2J7QRB7 A0A2T7NW75 A0A2J7PSI0 A0A2J7Q6T5 A0A2J7PSH5 A0A2J7QEY3
Pubmed
EMBL
GU815090
ADF18553.1
BABH01034706
FJ265548
ADI61816.1
KZ150025
+ More
PZC74827.1 LBMM01007694 KMQ89503.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 ODYU01006118 SOQ47682.1 DPCO01000033 HCE59365.1 FJ265557 ADI61825.1 FJ265553 ADI61821.1 FJ265552 ADI61820.1 FJ265547 ADI61815.1 JTDY01003909 KOB68843.1 FJ265560 ADI61828.1 LBMM01000801 KMQ97461.1 MRZV01001667 PIK36482.1 JTDY01001360 KOB74082.1 LBMM01011957 KMQ86519.1 KN538403 KHJ42451.1 KL367526 KFD66335.1 AMGL01013224 AMGL01013225 GBBM01006247 JAC29171.1 GBBM01003639 JAC31779.1 BDSA01000081 GBE63537.1 AFYH01196306 NEVH01025141 PNF15737.1 NEVH01024527 PNF17035.1 AAGJ04118689 AAGJ04161747 NEVH01022660 PNF18262.1 NEVH01022635 PNF18667.1 AFYH01136302 NEVH01000276 PNF43685.1 NEVH01023953 PNF17910.1 ABLF02040179 GGMS01012699 MBY81902.1 NEVH01005277 PNF39130.1 NEVH01006736 PNF36722.1 NEVH01003750 PNF40086.1 NEVH01014840 PNF27481.1 NEVH01018380 PNF23492.1 NEVH01017533 PNF24153.1 NEVH01015421 PNF26777.1 NEVH01002982 PNF41147.1 NEVH01021610 PNF19569.1 NEVH01007824 PNF35184.1 NEVH01021921 PNF19468.1 NEVH01011914 PNF31128.1 PZQS01000008 PVD25402.1 NEVH01021928 PNF19298.1 NEVH01017452 PNF24293.1 NEVH01021931 PNF19256.1 NEVH01015305 PNF27148.1
PZC74827.1 LBMM01007694 KMQ89503.1 KZ150203 PZC72267.1 FJ265559 ADI61827.1 ODYU01006118 SOQ47682.1 DPCO01000033 HCE59365.1 FJ265557 ADI61825.1 FJ265553 ADI61821.1 FJ265552 ADI61820.1 FJ265547 ADI61815.1 JTDY01003909 KOB68843.1 FJ265560 ADI61828.1 LBMM01000801 KMQ97461.1 MRZV01001667 PIK36482.1 JTDY01001360 KOB74082.1 LBMM01011957 KMQ86519.1 KN538403 KHJ42451.1 KL367526 KFD66335.1 AMGL01013224 AMGL01013225 GBBM01006247 JAC29171.1 GBBM01003639 JAC31779.1 BDSA01000081 GBE63537.1 AFYH01196306 NEVH01025141 PNF15737.1 NEVH01024527 PNF17035.1 AAGJ04118689 AAGJ04161747 NEVH01022660 PNF18262.1 NEVH01022635 PNF18667.1 AFYH01136302 NEVH01000276 PNF43685.1 NEVH01023953 PNF17910.1 ABLF02040179 GGMS01012699 MBY81902.1 NEVH01005277 PNF39130.1 NEVH01006736 PNF36722.1 NEVH01003750 PNF40086.1 NEVH01014840 PNF27481.1 NEVH01018380 PNF23492.1 NEVH01017533 PNF24153.1 NEVH01015421 PNF26777.1 NEVH01002982 PNF41147.1 NEVH01021610 PNF19569.1 NEVH01007824 PNF35184.1 NEVH01021921 PNF19468.1 NEVH01011914 PNF31128.1 PZQS01000008 PVD25402.1 NEVH01021928 PNF19298.1 NEVH01017452 PNF24293.1 NEVH01021931 PNF19256.1 NEVH01015305 PNF27148.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
D5LB39
H9JCQ5
D7F163
A0A2W1BNH4
A0A0J7KGF7
A0A2W1BGD9
+ More
D7F174 A0A2H1W4B8 A0A3D1L484 D7F172 D7F168 D7F167 D7F162 A0A0L7KZZ5 D7F175 A0A0J7L4A4 A0A2G8JL44 A0A0L7LF38 A0A0J7K8A5 A0A0B1PP52 A0A085NA39 W5P980 A0A023G4L3 A0A023GD30 A0A2H6KKM9 H3A4I6 A0A1S4EQZ4 A0A2J7PHB6 A0A2J7PL22 H3J6U9 A0A2J7PPK7 A0A2J7PQR6 H3A3N0 A0A2J7RS89 A0A2J7PNJ4 J9KN99 A0A2S2QXQ0 A0A2J7RE76 A0A2J7R7C2 A0A2J7RGX9 A0A2J7QFW5 A0A2J7Q4I0 A0A2J7Q6D5 A0A2J7QDX3 A0A2J7RJY3 A0A2J7PTB8 A0A2J7R2Y0 A0A2J7PT05 A0A2J7QRB7 A0A2T7NW75 A0A2J7PSI0 A0A2J7Q6T5 A0A2J7PSH5 A0A2J7QEY3
D7F174 A0A2H1W4B8 A0A3D1L484 D7F172 D7F168 D7F167 D7F162 A0A0L7KZZ5 D7F175 A0A0J7L4A4 A0A2G8JL44 A0A0L7LF38 A0A0J7K8A5 A0A0B1PP52 A0A085NA39 W5P980 A0A023G4L3 A0A023GD30 A0A2H6KKM9 H3A4I6 A0A1S4EQZ4 A0A2J7PHB6 A0A2J7PL22 H3J6U9 A0A2J7PPK7 A0A2J7PQR6 H3A3N0 A0A2J7RS89 A0A2J7PNJ4 J9KN99 A0A2S2QXQ0 A0A2J7RE76 A0A2J7R7C2 A0A2J7RGX9 A0A2J7QFW5 A0A2J7Q4I0 A0A2J7Q6D5 A0A2J7QDX3 A0A2J7RJY3 A0A2J7PTB8 A0A2J7R2Y0 A0A2J7PT05 A0A2J7QRB7 A0A2T7NW75 A0A2J7PSI0 A0A2J7Q6T5 A0A2J7PSH5 A0A2J7QEY3
Ontologies
PANTHER
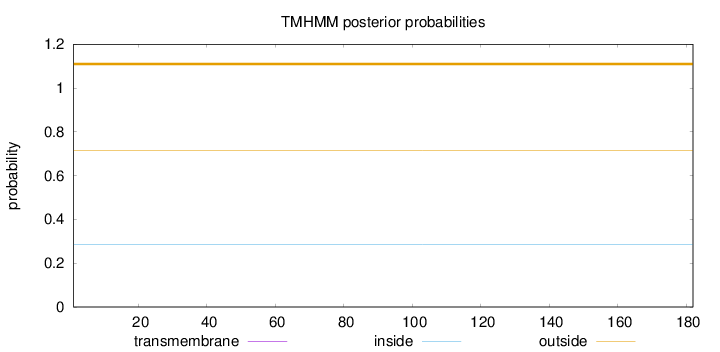
Topology
Length:
182
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.28396
outside
1 - 182
Population Genetic Test Statistics
Pi
0.260318
Theta
1.477877
Tajima's D
-1.567355
CLR
2.769384
CSRT
0.0395480225988701
Interpretation
Uncertain
