Gene
KWMTBOMO00284
Pre Gene Modal
BGIBMGA000547
Annotation
neuropeptide_receptor_B4_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.382
Sequence
CDS
ATGTTGTCACGTTTTCTGAACAAATTATTTAGTCAAACCAACGTCACTGAAAATCCACAGCTGAAGGTGCTTTATACACGTTATGAAGAGTGTTTTGTCTCAAATGAGACTCACAATACAATAGTTCAGTCGGAATACGACGAAAATGCTCCACGGTGCCCGAAAACGTTCGACGGTTTTTCATGTTGGGACGAAACTCCGTCCAACACAACGGTTGTACAAAATTGTCCAGAATTAATAAATGGATTCGACCCAAGCCGAACTGCTTTTAAAGAGTGTTTGGAGAACGGAACTTGGCTCGAGCATCCGGAGAGCAAGAAAGTATGGACCAATTACACGACATGCATTGACTACGATGATCTGCGTTTTCGCAACGTGATCAACAACATGTATGTCGGTGGCTACGCGATCTCATTGATCGCTCTGATCCTGTCTCTGATGATATTCGTTTTTCTCAGGCATTTTCAATATAAGCGGACGCGAATCCATGCCAATCTATTCATTTCGTTCATCCTCAACAACATCATGTGGATCGTCTGGTATAAAACTGTTGTGGACTACATCGAAGTGGTACAAGAAAACACGATTTGGTGTCAATCCCTTCACATCTTGACCTATTATCACATGATGACGAGCTACATGTGGATGTTCTGCGAAGGCCTACACCTGCATGTTGGACTGGTGTCTATAAATGAATATATCTCAGTACGCTCATACTGTGCCATCGGGTGGGCCATCCCAGCTCTGGTTGTCGCTCTATATACTGGTGTTCGTATGCAACTCCGTTATGGAACAGAACGATGCTGGATGGATCAAAGCCACGCTCTCTGGATCATCGTCATCGCAGTTGTGATTATACTCACACTTTCCTTCCTATTTCTGGTGAACGTTATTCGAGTTCTGTTAAAAAAGATGCAACCGCCCGCCTCAAATCAGCCGAATGAAGCCGCCAAGAAAGCTGCCCGGGCCACTGCCACTTTGGTTATATATTTCATGATTCCGCTGTACGGACTGCATTTCATCTTGATCCCATTTCAACGAGCTCCGGAATCTATTGGCGAGAAAATATACCAAGTTGTTTCAGCGTTGCTCACAAGCTTGCAGGGACTCTGTGTTTCTATTTTGTTTTGTTTTACAAATAACGACGTAAAGACAGCCCTTACAAATTACTTGGCACGGTACAAGCGAAAAACCGAAGCTATCCAGATGACTGGTATCACCACTGGAGAAAGCGCTGTGAATCCAAATGCCGCCTAG
Protein
MLSRFLNKLFSQTNVTENPQLKVLYTRYEECFVSNETHNTIVQSEYDENAPRCPKTFDGFSCWDETPSNTTVVQNCPELINGFDPSRTAFKECLENGTWLEHPESKKVWTNYTTCIDYDDLRFRNVINNMYVGGYAISLIALILSLMIFVFLRHFQYKRTRIHANLFISFILNNIMWIVWYKTVVDYIEVVQENTIWCQSLHILTYYHMMTSYMWMFCEGLHLHVGLVSINEYISVRSYCAIGWAIPALVVALYTGVRMQLRYGTERCWMDQSHALWIIVIAVVIILTLSFLFLVNVIRVLLKKMQPPASNQPNEAAKKAARATATLVIYFMIPLYGLHFILIPFQRAPESIGEKIYQVVSALLTSLQGLCVSILFCFTNNDVKTALTNYLARYKRKTEAIQMTGITTGESAVNPNAA
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
B3XXQ2
A0A0S1YDB6
A0A2A4JRA7
A0A212ETG2
A0A2A4JS52
A0A2A4JRS8
+ More
U3U988 V5V0N7 A0A2H8TY46 W5JX54 A0A182UZR5 A0A182Q2A5 A0A2U9PFX0 A0A182XP26 A0A182HK63 A0A2J7PNW2 A0A182RD90 A0A182FS81 A0A182VW55 A0A1L8DK21 A0A336KK11 A0A182NQW8 A0A1B0GHH2 A0A1J1J1H9 A0A2S2QIM3 A0A067R1M3 E9HF82 A0A182K5B5 T1KMN8 D6WM84 A0A1W4X1S2 A0A1I8PM98 A0A0A9XCA8 A0A1B6ECT6 A0A1B6E976 A0A2P8XJ43 A0A345BTT1 A0A0K8VE06 A0A0K8U007 A0A1I8PM52 A0A3B0KVK5 A0A034W1B7 Q29FU0 A0A2U9PFW0 A0A1I8ML57 T1HL11 B4NEI4 A0A0Q9XEN2 B4L605 A0A1W4V6Y5 B4Q1Z1 A0A0L0C3J0 V5UZ27 A0A1A9YQG6 V5UZV0 B3NX83 U3UA10 A0A1B6GBP0 A0A1B6FEL9 Q9VYH9 A0A1B0G808 A0A1B6HNS4 A0A1B6LXJ3 T1JAY3 B4IGK9 N6U705 U4UHU1 A0A1B0A198 K7J018 B3MS26 A0A2S2PE39 A0A3Q0IXJ0 A0A3L8DW89 A0A336MHY5 A0A224YUH5 U3U4H7 X1WIB5 A0A131YYY3 B4M747 A0A0C9RV18 A0A088A7A3 A0A182PRA0 A0A1L8DJN0 A0A1L8DJZ5 A0A1L8DJU9 A0A1B6DAB7 B4JM00 A0A1W4XSW4 E0VT86 A0A067R9A4 A0A2R7WD56 A0A0K8VAV3 A0A2S2NU23 A0A0K8W8K9
U3U988 V5V0N7 A0A2H8TY46 W5JX54 A0A182UZR5 A0A182Q2A5 A0A2U9PFX0 A0A182XP26 A0A182HK63 A0A2J7PNW2 A0A182RD90 A0A182FS81 A0A182VW55 A0A1L8DK21 A0A336KK11 A0A182NQW8 A0A1B0GHH2 A0A1J1J1H9 A0A2S2QIM3 A0A067R1M3 E9HF82 A0A182K5B5 T1KMN8 D6WM84 A0A1W4X1S2 A0A1I8PM98 A0A0A9XCA8 A0A1B6ECT6 A0A1B6E976 A0A2P8XJ43 A0A345BTT1 A0A0K8VE06 A0A0K8U007 A0A1I8PM52 A0A3B0KVK5 A0A034W1B7 Q29FU0 A0A2U9PFW0 A0A1I8ML57 T1HL11 B4NEI4 A0A0Q9XEN2 B4L605 A0A1W4V6Y5 B4Q1Z1 A0A0L0C3J0 V5UZ27 A0A1A9YQG6 V5UZV0 B3NX83 U3UA10 A0A1B6GBP0 A0A1B6FEL9 Q9VYH9 A0A1B0G808 A0A1B6HNS4 A0A1B6LXJ3 T1JAY3 B4IGK9 N6U705 U4UHU1 A0A1B0A198 K7J018 B3MS26 A0A2S2PE39 A0A3Q0IXJ0 A0A3L8DW89 A0A336MHY5 A0A224YUH5 U3U4H7 X1WIB5 A0A131YYY3 B4M747 A0A0C9RV18 A0A088A7A3 A0A182PRA0 A0A1L8DJN0 A0A1L8DJZ5 A0A1L8DJU9 A0A1B6DAB7 B4JM00 A0A1W4XSW4 E0VT86 A0A067R9A4 A0A2R7WD56 A0A0K8VAV3 A0A2S2NU23 A0A0K8W8K9
Pubmed
18725956
22118469
23932938
24312424
20920257
23761445
+ More
24845553 21292972 18362917 19820115 25401762 26823975 29403074 30022930 25348373 15632085 17994087 23185243 25315136 18057021 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 20075255 30249741 28797301 26830274 20566863
24845553 21292972 18362917 19820115 25401762 26823975 29403074 30022930 25348373 15632085 17994087 23185243 25315136 18057021 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23537049 20075255 30249741 28797301 26830274 20566863
EMBL
AB330460
BAG68438.1
KT031046
RSAL01000118
ALM88344.1
RVE46782.1
+ More
NWSH01000732 PCG74521.1 AGBW02012593 OWR44767.1 PCG74518.1 PCG74519.1 AB817337 BAO01104.1 KF446640 AHB86571.1 GFXV01006916 MBW18721.1 ADMH02000021 ETN68065.1 AXCN02001488 MG550230 AWT50664.1 APCN01000817 NEVH01023284 PNF18006.1 GFDF01007390 JAV06694.1 UFQS01000602 UFQT01000602 SSX05266.1 SSX25627.1 AJWK01004622 CVRI01000064 CRL05620.1 GGMS01008157 MBY77360.1 KK853094 KDR11499.1 GL732634 EFX69566.1 CAEY01000248 KQ971343 EFA04592.1 GBHO01028889 GDHC01017884 JAG14715.1 JAQ00745.1 GEDC01001553 JAS35745.1 GEDC01002853 JAS34445.1 PYGN01001953 PSN32019.1 MH331892 AXF54360.1 GDHF01015564 JAI36750.1 GDHF01032240 JAI20074.1 OUUW01000021 SPP89391.1 GAKP01009601 JAC49351.1 CH379065 EAL31489.3 KRT07222.1 KRT07223.1 KRT07224.1 MG550228 AWT50662.1 ACPB03005233 CH964239 EDW82153.1 CH933812 KRG07040.1 EDW05801.1 KRG07041.1 CM000162 EDX01512.2 KRK06192.1 JRES01000955 KNC26817.1 KC660149 AHB86317.1 KC660150 AHB86318.1 CH954180 EDV47255.2 AB817336 BAO01103.1 GECZ01010056 JAS59713.1 GECZ01021215 JAS48554.1 AE014298 AAF48216.2 AHN59644.1 CCAG010000149 GECU01031372 JAS76334.1 GEBQ01025341 GEBQ01011586 JAT14636.1 JAT28391.1 JH432007 CH480836 EDW48959.1 APGK01037673 APGK01037674 APGK01037675 KB740948 ENN77430.1 KB632364 ERL93564.1 CH902622 EDV34581.2 GGMR01015101 MBY27720.1 QOIP01000003 RLU24626.1 SSX05267.1 SSX25628.1 GFPF01006478 MAA17624.1 AB817334 BAO01101.1 ABLF02019819 ABLF02019820 ABLF02019822 GEDV01004163 JAP84394.1 CH940653 EDW62614.2 GBYB01011516 JAG81283.1 GFDF01007421 JAV06663.1 GFDF01007420 JAV06664.1 GFDF01007346 JAV06738.1 GEDC01014657 JAS22641.1 CH916371 EDV91761.1 DS235760 EEB16592.1 KK852804 KDR16174.1 KK854645 PTY17597.1 GDHF01016338 JAI35976.1 GGMR01008082 MBY20701.1 GDHF01004955 JAI47359.1
NWSH01000732 PCG74521.1 AGBW02012593 OWR44767.1 PCG74518.1 PCG74519.1 AB817337 BAO01104.1 KF446640 AHB86571.1 GFXV01006916 MBW18721.1 ADMH02000021 ETN68065.1 AXCN02001488 MG550230 AWT50664.1 APCN01000817 NEVH01023284 PNF18006.1 GFDF01007390 JAV06694.1 UFQS01000602 UFQT01000602 SSX05266.1 SSX25627.1 AJWK01004622 CVRI01000064 CRL05620.1 GGMS01008157 MBY77360.1 KK853094 KDR11499.1 GL732634 EFX69566.1 CAEY01000248 KQ971343 EFA04592.1 GBHO01028889 GDHC01017884 JAG14715.1 JAQ00745.1 GEDC01001553 JAS35745.1 GEDC01002853 JAS34445.1 PYGN01001953 PSN32019.1 MH331892 AXF54360.1 GDHF01015564 JAI36750.1 GDHF01032240 JAI20074.1 OUUW01000021 SPP89391.1 GAKP01009601 JAC49351.1 CH379065 EAL31489.3 KRT07222.1 KRT07223.1 KRT07224.1 MG550228 AWT50662.1 ACPB03005233 CH964239 EDW82153.1 CH933812 KRG07040.1 EDW05801.1 KRG07041.1 CM000162 EDX01512.2 KRK06192.1 JRES01000955 KNC26817.1 KC660149 AHB86317.1 KC660150 AHB86318.1 CH954180 EDV47255.2 AB817336 BAO01103.1 GECZ01010056 JAS59713.1 GECZ01021215 JAS48554.1 AE014298 AAF48216.2 AHN59644.1 CCAG010000149 GECU01031372 JAS76334.1 GEBQ01025341 GEBQ01011586 JAT14636.1 JAT28391.1 JH432007 CH480836 EDW48959.1 APGK01037673 APGK01037674 APGK01037675 KB740948 ENN77430.1 KB632364 ERL93564.1 CH902622 EDV34581.2 GGMR01015101 MBY27720.1 QOIP01000003 RLU24626.1 SSX05267.1 SSX25628.1 GFPF01006478 MAA17624.1 AB817334 BAO01101.1 ABLF02019819 ABLF02019820 ABLF02019822 GEDV01004163 JAP84394.1 CH940653 EDW62614.2 GBYB01011516 JAG81283.1 GFDF01007421 JAV06663.1 GFDF01007420 JAV06664.1 GFDF01007346 JAV06738.1 GEDC01014657 JAS22641.1 CH916371 EDV91761.1 DS235760 EEB16592.1 KK852804 KDR16174.1 KK854645 PTY17597.1 GDHF01016338 JAI35976.1 GGMR01008082 MBY20701.1 GDHF01004955 JAI47359.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000000673
UP000075903
UP000075886
+ More
UP000076407 UP000075840 UP000235965 UP000075900 UP000069272 UP000075920 UP000075884 UP000092461 UP000183832 UP000027135 UP000000305 UP000075881 UP000015104 UP000007266 UP000192223 UP000095300 UP000245037 UP000268350 UP000001819 UP000095301 UP000015103 UP000007798 UP000009192 UP000192221 UP000002282 UP000037069 UP000092443 UP000008711 UP000000803 UP000092444 UP000001292 UP000019118 UP000030742 UP000092445 UP000002358 UP000007801 UP000079169 UP000279307 UP000007819 UP000008792 UP000005203 UP000075885 UP000001070 UP000009046
UP000076407 UP000075840 UP000235965 UP000075900 UP000069272 UP000075920 UP000075884 UP000092461 UP000183832 UP000027135 UP000000305 UP000075881 UP000015104 UP000007266 UP000192223 UP000095300 UP000245037 UP000268350 UP000001819 UP000095301 UP000015103 UP000007798 UP000009192 UP000192221 UP000002282 UP000037069 UP000092443 UP000008711 UP000000803 UP000092444 UP000001292 UP000019118 UP000030742 UP000092445 UP000002358 UP000007801 UP000079169 UP000279307 UP000007819 UP000008792 UP000005203 UP000075885 UP000001070 UP000009046
Interpro
Gene 3D
ProteinModelPortal
B3XXQ2
A0A0S1YDB6
A0A2A4JRA7
A0A212ETG2
A0A2A4JS52
A0A2A4JRS8
+ More
U3U988 V5V0N7 A0A2H8TY46 W5JX54 A0A182UZR5 A0A182Q2A5 A0A2U9PFX0 A0A182XP26 A0A182HK63 A0A2J7PNW2 A0A182RD90 A0A182FS81 A0A182VW55 A0A1L8DK21 A0A336KK11 A0A182NQW8 A0A1B0GHH2 A0A1J1J1H9 A0A2S2QIM3 A0A067R1M3 E9HF82 A0A182K5B5 T1KMN8 D6WM84 A0A1W4X1S2 A0A1I8PM98 A0A0A9XCA8 A0A1B6ECT6 A0A1B6E976 A0A2P8XJ43 A0A345BTT1 A0A0K8VE06 A0A0K8U007 A0A1I8PM52 A0A3B0KVK5 A0A034W1B7 Q29FU0 A0A2U9PFW0 A0A1I8ML57 T1HL11 B4NEI4 A0A0Q9XEN2 B4L605 A0A1W4V6Y5 B4Q1Z1 A0A0L0C3J0 V5UZ27 A0A1A9YQG6 V5UZV0 B3NX83 U3UA10 A0A1B6GBP0 A0A1B6FEL9 Q9VYH9 A0A1B0G808 A0A1B6HNS4 A0A1B6LXJ3 T1JAY3 B4IGK9 N6U705 U4UHU1 A0A1B0A198 K7J018 B3MS26 A0A2S2PE39 A0A3Q0IXJ0 A0A3L8DW89 A0A336MHY5 A0A224YUH5 U3U4H7 X1WIB5 A0A131YYY3 B4M747 A0A0C9RV18 A0A088A7A3 A0A182PRA0 A0A1L8DJN0 A0A1L8DJZ5 A0A1L8DJU9 A0A1B6DAB7 B4JM00 A0A1W4XSW4 E0VT86 A0A067R9A4 A0A2R7WD56 A0A0K8VAV3 A0A2S2NU23 A0A0K8W8K9
U3U988 V5V0N7 A0A2H8TY46 W5JX54 A0A182UZR5 A0A182Q2A5 A0A2U9PFX0 A0A182XP26 A0A182HK63 A0A2J7PNW2 A0A182RD90 A0A182FS81 A0A182VW55 A0A1L8DK21 A0A336KK11 A0A182NQW8 A0A1B0GHH2 A0A1J1J1H9 A0A2S2QIM3 A0A067R1M3 E9HF82 A0A182K5B5 T1KMN8 D6WM84 A0A1W4X1S2 A0A1I8PM98 A0A0A9XCA8 A0A1B6ECT6 A0A1B6E976 A0A2P8XJ43 A0A345BTT1 A0A0K8VE06 A0A0K8U007 A0A1I8PM52 A0A3B0KVK5 A0A034W1B7 Q29FU0 A0A2U9PFW0 A0A1I8ML57 T1HL11 B4NEI4 A0A0Q9XEN2 B4L605 A0A1W4V6Y5 B4Q1Z1 A0A0L0C3J0 V5UZ27 A0A1A9YQG6 V5UZV0 B3NX83 U3UA10 A0A1B6GBP0 A0A1B6FEL9 Q9VYH9 A0A1B0G808 A0A1B6HNS4 A0A1B6LXJ3 T1JAY3 B4IGK9 N6U705 U4UHU1 A0A1B0A198 K7J018 B3MS26 A0A2S2PE39 A0A3Q0IXJ0 A0A3L8DW89 A0A336MHY5 A0A224YUH5 U3U4H7 X1WIB5 A0A131YYY3 B4M747 A0A0C9RV18 A0A088A7A3 A0A182PRA0 A0A1L8DJN0 A0A1L8DJZ5 A0A1L8DJU9 A0A1B6DAB7 B4JM00 A0A1W4XSW4 E0VT86 A0A067R9A4 A0A2R7WD56 A0A0K8VAV3 A0A2S2NU23 A0A0K8W8K9
PDB
5UZ7
E-value=2.80237e-51,
Score=510
Ontologies
GO
Topology
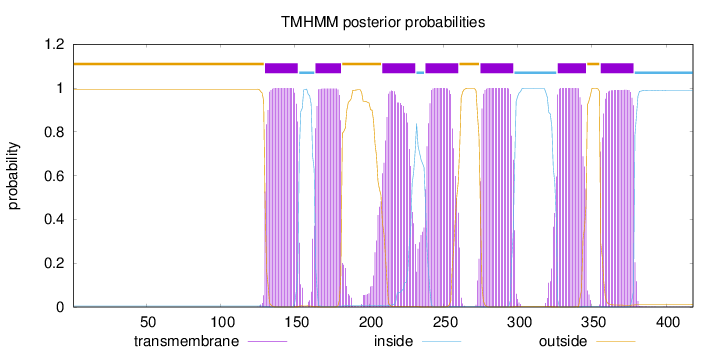
Length:
418
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
150.68267
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00501
outside
1 - 129
TMhelix
130 - 152
inside
153 - 163
TMhelix
164 - 181
outside
182 - 208
TMhelix
209 - 231
inside
232 - 237
TMhelix
238 - 260
outside
261 - 274
TMhelix
275 - 297
inside
298 - 326
TMhelix
327 - 346
outside
347 - 355
TMhelix
356 - 378
inside
379 - 418
Population Genetic Test Statistics
Pi
6.040058
Theta
14.209106
Tajima's D
-1.087129
CLR
0.138119
CSRT
0.119394030298485
Interpretation
Uncertain
