Gene
KWMTBOMO00283
Pre Gene Modal
BGIBMGA000675
Annotation
PREDICTED:_sodium/potassium/calcium_exchanger_3-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.95
Sequence
CDS
ATGGAACAAATTAAGGTTGCAAATCTAGCTGTCTTAGTGGGTTTGGTTGCTAGTTTTTATACTGTATCAGCCATCGAGGATTTCAGTTTGTCGAACGACTCTGTTTATTTACGTGTGGAGAGGAATGGGAAAATGATGTACATGACAGAAGAACTAATAAATAAATTAGAACTAGAAGATACGACTTGGCATTGGAAACCATGGCTGAGGACCAGAATTGAAGTCGTTGATAACGTTGCACATGAAGTGGTCTGCGCAACAGGAGATTCCATAGACAGTTTTCCTGATGGTATATTTACAGATGAACAACTCCGTCAAGGTGCCTTCCTTTTGTATGTCGTCTTTGGCATTTACACATTCACACTTTTGGCTATTGTTTGCAATGATTATTTCATTCCTTGTGTTGAACTTATTTGCGAAGACTTGAAGATCCCACAAAACGTAGCAGCAGCTACATTTATGTCTGTCGCGACGTCTTGTCCAGAATTCTTCGTGAACGTGATATCAACCTTTCTAACTGAATCGGATATGGGTATCGGAACTATTGTAGGATCCGCTCTTTTTAATGCTCTCGGAGTAGCTGCTATTGGAGGCCTTGCTGCTATTACACCAATAGCTATCGACATGCGGCCAGTGACAAGAGATGTGCTAATATACATGGTAAATATTAGCGTTCTTGTGGCATTTGTATGGGACGGCCAAATCGATTGGTTTGAGGCGATGGTTCTAGGAATTCTCTATATTTTGTATTTCGTTGTCATGTTTAACAGTATGAAAGCGTTTGTTCTTATTGATAAAATTTGGGCAGCCTGTTGCAAAAAGAATAATGGAGTTACCGCATCTTTGAACGATAACAATGTGTCCGTGGAAGAGGGTATTGATAACAAAGGTTTCAACGACCCCAGTCCTGTTACGAATGGAACCACTATGACGGAGAAGCCAGCTATTGATGAAAAAAATGATGTTGAAGAAACGCTAAATAAAAAAAGTTTGTTTACTTTTCCTTTTCACAAATCAATACTATACAAGATATGGTGGTTTTACATTTGGCCTTTGAAGCTGGTGCTTGGGTTGACTATACCCTCTCCAGTTACTCGAAAAAAGTTATATCCTCTCGCTTTCATAATGTGCATCGTGTGGATTGGAGTGAATTCATACTTTGTGTCTTGGAGTATGACTGTGATAGGACATACCTTCTTTATTCCTGAATCAGTTATGGGGATGACATTTTTAGCATTTGGAGGATGTCTACCAGAAGCGTGCTCGATATTTATTATGTCTAGAAGAGGCGAAGGTGGTATTGGGGTCTCAAATGCACTTGGAGCCAACTCCTTAGCGATTTTGTTTGCTTTGGGCTTACCGTGGTTGATAAGGACAGTTTCGCTGGAAATTCAAGGAGCTGAGAATACCGCAGTACTAATAAACTCAGCAGGTATCGACTTTGTCGTGGGATCTCTTCTTGTAGCTGTTGTCAGTTTATGGATCGTCCTGTTCATTGGAAAATTCACGCTGAGACGTACTGTAGGTATAGTGCTAGGGATACTTTATGTCGTTTTTATCGTTTTCGCTATTCTCGTGGAGTTGGGAATCATTTTCGATCGAAACGTTGAGTTGAGCTCGTGCTGA
Protein
MEQIKVANLAVLVGLVASFYTVSAIEDFSLSNDSVYLRVERNGKMMYMTEELINKLELEDTTWHWKPWLRTRIEVVDNVAHEVVCATGDSIDSFPDGIFTDEQLRQGAFLLYVVFGIYTFTLLAIVCNDYFIPCVELICEDLKIPQNVAAATFMSVATSCPEFFVNVISTFLTESDMGIGTIVGSALFNALGVAAIGGLAAITPIAIDMRPVTRDVLIYMVNISVLVAFVWDGQIDWFEAMVLGILYILYFVVMFNSMKAFVLIDKIWAACCKKNNGVTASLNDNNVSVEEGIDNKGFNDPSPVTNGTTMTEKPAIDEKNDVEETLNKKSLFTFPFHKSILYKIWWFYIWPLKLVLGLTIPSPVTRKKLYPLAFIMCIVWIGVNSYFVSWSMTVIGHTFFIPESVMGMTFLAFGGCLPEACSIFIMSRRGEGGIGVSNALGANSLAILFALGLPWLIRTVSLEIQGAENTAVLINSAGIDFVVGSLLVAVVSLWIVLFIGKFTLRRTVGIVLGILYVVFIVFAILVELGIIFDRNVELSSC
Summary
Similarity
Belongs to the Ca(2+):cation antiporter (CaCA) (TC 2.A.19) family.
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
A0A2A4JT07
A0A2H1WI79
A0A194QVA8
A0A194PTP8
A0A212ETE8
A0A084V9V9
+ More
A0A1Y9HFE4 A0A182Q635 A0A1S4F8Z8 Q17BT5 A0A182GKT4 A0A182VNA7 A0A182U315 A0A182MRU6 A0A2C9GR81 A0A182KKG1 A0A1L8DK38 W5JWA6 A0A182Y9M0 A0A182WUK8 A0A182J077 A0A182WE41 V5I898 A0A182FV54 A0A182N2P2 A0A1B0GMI3 A0A182PDA0 A0A084V9W0 A0A182K8I5 Q7PSJ7 A0A1B0GMI4 A0A2C9GR82 Q17BT4 A0A182JHY5 A0A1S4F8P6 D6WM87 A0A182Q6T2 W5JU26 A0A182UQI0 A0A2A3E1A4 A0A1W7R6T4 A0A088AGL8 A0A0T6AXF0 U4UKB9 A0A1I8MJH1 U4UTP4 A0A1B6D9C7 A0A0K8TR74 A0A034W6P0 A0A067R056 B0WBJ4 A0A067RBZ5 A0A1B6M4M0 A0A0K8US07 N6UFJ8 N6THY4 W8BQP8 A0A0K8W873 W8C8K6 A0A0Q9W6E5 A0A0L0BYT5 E2C4F0 A0A310S7P6 A0A0A9XWU9 A0A0A1XSB5 A0A224XMZ1 N6US54 A0A0M9A307 A0A0M3QZN1 A0A023EZI4 A0A2J7QAG0 A0A1W4W1Z0 A0A1W4W1Y6 A0A0L7RBA1 A0A1B6FZ48 A0A1W4WDX3 A0A1B6HL49 A0A021WWX0 A0A1B6JE47 B3MYI6 B4IM40 A0A1W4W1Z1 A0A336LEQ5 A0A1W4WCL5 A0A0J9R6C9 Q7PLW4 A0A1B6KAW2 A0A0N8NZE4 A0A0Q9XHC3 A0A1A9W8U2 A0A0J9R7S8 Q5LJY0 A0A0J9R6Q2 A0A0J9TUA0 A0A1A9XBW7 A0A0J9R6L8 A0A1I8NQJ2 A0A1I8NQJ4 A0A088AGL0
A0A1Y9HFE4 A0A182Q635 A0A1S4F8Z8 Q17BT5 A0A182GKT4 A0A182VNA7 A0A182U315 A0A182MRU6 A0A2C9GR81 A0A182KKG1 A0A1L8DK38 W5JWA6 A0A182Y9M0 A0A182WUK8 A0A182J077 A0A182WE41 V5I898 A0A182FV54 A0A182N2P2 A0A1B0GMI3 A0A182PDA0 A0A084V9W0 A0A182K8I5 Q7PSJ7 A0A1B0GMI4 A0A2C9GR82 Q17BT4 A0A182JHY5 A0A1S4F8P6 D6WM87 A0A182Q6T2 W5JU26 A0A182UQI0 A0A2A3E1A4 A0A1W7R6T4 A0A088AGL8 A0A0T6AXF0 U4UKB9 A0A1I8MJH1 U4UTP4 A0A1B6D9C7 A0A0K8TR74 A0A034W6P0 A0A067R056 B0WBJ4 A0A067RBZ5 A0A1B6M4M0 A0A0K8US07 N6UFJ8 N6THY4 W8BQP8 A0A0K8W873 W8C8K6 A0A0Q9W6E5 A0A0L0BYT5 E2C4F0 A0A310S7P6 A0A0A9XWU9 A0A0A1XSB5 A0A224XMZ1 N6US54 A0A0M9A307 A0A0M3QZN1 A0A023EZI4 A0A2J7QAG0 A0A1W4W1Z0 A0A1W4W1Y6 A0A0L7RBA1 A0A1B6FZ48 A0A1W4WDX3 A0A1B6HL49 A0A021WWX0 A0A1B6JE47 B3MYI6 B4IM40 A0A1W4W1Z1 A0A336LEQ5 A0A1W4WCL5 A0A0J9R6C9 Q7PLW4 A0A1B6KAW2 A0A0N8NZE4 A0A0Q9XHC3 A0A1A9W8U2 A0A0J9R7S8 Q5LJY0 A0A0J9R6Q2 A0A0J9TUA0 A0A1A9XBW7 A0A0J9R6L8 A0A1I8NQJ2 A0A1I8NQJ4 A0A088AGL0
Pubmed
26354079
22118469
24438588
17510324
26483478
20966253
+ More
20920257 23761445 25244985 12364791 18362917 19820115 23537049 25315136 26369729 25348373 24845553 24495485 17994087 26108605 20798317 25401762 26823975 25830018 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356
20920257 23761445 25244985 12364791 18362917 19820115 23537049 25315136 26369729 25348373 24845553 24495485 17994087 26108605 20798317 25401762 26823975 25830018 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356
EMBL
NWSH01000732
PCG74522.1
ODYU01008830
SOQ52775.1
KQ461108
KPJ09254.1
+ More
KQ459594 KPI96129.1 AGBW02012593 OWR44766.1 ATLV01000460 KE523882 KFB34753.1 AXCN02001080 CH477316 EAT43778.1 JXUM01070739 JXUM01070740 JXUM01070741 JXUM01070742 JXUM01070743 KQ562624 KXJ75461.1 AXCM01007828 APCN01001433 GFDF01007347 JAV06737.1 ADMH02000090 ETN67853.1 GALX01005060 JAB63406.1 AJVK01011474 AJVK01011475 KFB34754.1 AAAB01008823 EAA05479.5 AJVK01011476 EAT43779.1 KQ971343 EFA03343.2 ETN67852.1 KZ288460 PBC25477.1 GEHC01000815 JAV46830.1 LJIG01022574 KRT79859.1 KB632364 ERL93567.1 ERL93566.1 GEDC01015015 JAS22283.1 GDAI01000724 JAI16879.1 GAKP01008569 GAKP01008568 JAC50384.1 KK852804 KDR16220.1 DS231879 EDS42433.1 KDR16218.1 GEBQ01009123 JAT30854.1 GDHF01024865 GDHF01023184 JAI27449.1 JAI29130.1 APGK01037673 KB740948 ENN77427.1 ENN77428.1 GAMC01002975 JAC03581.1 GDHF01004911 JAI47403.1 GAMC01002974 JAC03582.1 CH940653 KRF80531.1 JRES01001143 KNC25185.1 GL452462 EFN77187.1 KQ776822 OAD52169.1 GBHO01020256 GBRD01011629 GDHC01019424 JAG23348.1 JAG54195.1 JAP99204.1 GBXI01000396 JAD13896.1 GFTR01006474 JAW09952.1 APGK01018911 KB740085 KB632303 ENN81547.1 ERL91812.1 KQ435752 KOX76187.1 CP012528 ALC49669.1 GBBI01004059 JAC14653.1 NEVH01016330 PNF25571.1 KQ414617 KOC68021.1 GECZ01024111 GECZ01014292 JAS45658.1 JAS55477.1 GECU01032286 JAS75420.1 AE014298 EYR77335.1 GECU01010473 JAS97233.1 CH902632 EDV32680.2 CH480924 EDW43676.1 UFQS01001835 UFQT01001835 SSX12512.1 SSX31955.1 CM002911 KMY91737.1 BT125892 ADV19033.1 EAA46196.2 GEBQ01031390 GEBQ01026499 GEBQ01018163 GEBQ01007137 JAT08587.1 JAT13478.1 JAT21814.1 JAT32840.1 KPU74092.1 CH933812 KRG07049.1 KMY91739.1 BT099540 ACU32624.1 EAL24595.2 KMY91736.1 KMY91740.1 KMY91738.1
KQ459594 KPI96129.1 AGBW02012593 OWR44766.1 ATLV01000460 KE523882 KFB34753.1 AXCN02001080 CH477316 EAT43778.1 JXUM01070739 JXUM01070740 JXUM01070741 JXUM01070742 JXUM01070743 KQ562624 KXJ75461.1 AXCM01007828 APCN01001433 GFDF01007347 JAV06737.1 ADMH02000090 ETN67853.1 GALX01005060 JAB63406.1 AJVK01011474 AJVK01011475 KFB34754.1 AAAB01008823 EAA05479.5 AJVK01011476 EAT43779.1 KQ971343 EFA03343.2 ETN67852.1 KZ288460 PBC25477.1 GEHC01000815 JAV46830.1 LJIG01022574 KRT79859.1 KB632364 ERL93567.1 ERL93566.1 GEDC01015015 JAS22283.1 GDAI01000724 JAI16879.1 GAKP01008569 GAKP01008568 JAC50384.1 KK852804 KDR16220.1 DS231879 EDS42433.1 KDR16218.1 GEBQ01009123 JAT30854.1 GDHF01024865 GDHF01023184 JAI27449.1 JAI29130.1 APGK01037673 KB740948 ENN77427.1 ENN77428.1 GAMC01002975 JAC03581.1 GDHF01004911 JAI47403.1 GAMC01002974 JAC03582.1 CH940653 KRF80531.1 JRES01001143 KNC25185.1 GL452462 EFN77187.1 KQ776822 OAD52169.1 GBHO01020256 GBRD01011629 GDHC01019424 JAG23348.1 JAG54195.1 JAP99204.1 GBXI01000396 JAD13896.1 GFTR01006474 JAW09952.1 APGK01018911 KB740085 KB632303 ENN81547.1 ERL91812.1 KQ435752 KOX76187.1 CP012528 ALC49669.1 GBBI01004059 JAC14653.1 NEVH01016330 PNF25571.1 KQ414617 KOC68021.1 GECZ01024111 GECZ01014292 JAS45658.1 JAS55477.1 GECU01032286 JAS75420.1 AE014298 EYR77335.1 GECU01010473 JAS97233.1 CH902632 EDV32680.2 CH480924 EDW43676.1 UFQS01001835 UFQT01001835 SSX12512.1 SSX31955.1 CM002911 KMY91737.1 BT125892 ADV19033.1 EAA46196.2 GEBQ01031390 GEBQ01026499 GEBQ01018163 GEBQ01007137 JAT08587.1 JAT13478.1 JAT21814.1 JAT32840.1 KPU74092.1 CH933812 KRG07049.1 KMY91739.1 BT099540 ACU32624.1 EAL24595.2 KMY91736.1 KMY91740.1 KMY91738.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000030765
UP000075900
+ More
UP000075886 UP000008820 UP000069940 UP000249989 UP000075903 UP000075902 UP000075883 UP000075840 UP000075882 UP000000673 UP000076408 UP000076407 UP000075880 UP000075920 UP000069272 UP000075884 UP000092462 UP000075885 UP000075881 UP000007062 UP000007266 UP000242457 UP000005203 UP000030742 UP000095301 UP000027135 UP000002320 UP000019118 UP000008792 UP000037069 UP000008237 UP000053105 UP000092553 UP000235965 UP000192221 UP000053825 UP000000803 UP000007801 UP000001292 UP000009192 UP000091820 UP000092443 UP000095300
UP000075886 UP000008820 UP000069940 UP000249989 UP000075903 UP000075902 UP000075883 UP000075840 UP000075882 UP000000673 UP000076408 UP000076407 UP000075880 UP000075920 UP000069272 UP000075884 UP000092462 UP000075885 UP000075881 UP000007062 UP000007266 UP000242457 UP000005203 UP000030742 UP000095301 UP000027135 UP000002320 UP000019118 UP000008792 UP000037069 UP000008237 UP000053105 UP000092553 UP000235965 UP000192221 UP000053825 UP000000803 UP000007801 UP000001292 UP000009192 UP000091820 UP000092443 UP000095300
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4JT07
A0A2H1WI79
A0A194QVA8
A0A194PTP8
A0A212ETE8
A0A084V9V9
+ More
A0A1Y9HFE4 A0A182Q635 A0A1S4F8Z8 Q17BT5 A0A182GKT4 A0A182VNA7 A0A182U315 A0A182MRU6 A0A2C9GR81 A0A182KKG1 A0A1L8DK38 W5JWA6 A0A182Y9M0 A0A182WUK8 A0A182J077 A0A182WE41 V5I898 A0A182FV54 A0A182N2P2 A0A1B0GMI3 A0A182PDA0 A0A084V9W0 A0A182K8I5 Q7PSJ7 A0A1B0GMI4 A0A2C9GR82 Q17BT4 A0A182JHY5 A0A1S4F8P6 D6WM87 A0A182Q6T2 W5JU26 A0A182UQI0 A0A2A3E1A4 A0A1W7R6T4 A0A088AGL8 A0A0T6AXF0 U4UKB9 A0A1I8MJH1 U4UTP4 A0A1B6D9C7 A0A0K8TR74 A0A034W6P0 A0A067R056 B0WBJ4 A0A067RBZ5 A0A1B6M4M0 A0A0K8US07 N6UFJ8 N6THY4 W8BQP8 A0A0K8W873 W8C8K6 A0A0Q9W6E5 A0A0L0BYT5 E2C4F0 A0A310S7P6 A0A0A9XWU9 A0A0A1XSB5 A0A224XMZ1 N6US54 A0A0M9A307 A0A0M3QZN1 A0A023EZI4 A0A2J7QAG0 A0A1W4W1Z0 A0A1W4W1Y6 A0A0L7RBA1 A0A1B6FZ48 A0A1W4WDX3 A0A1B6HL49 A0A021WWX0 A0A1B6JE47 B3MYI6 B4IM40 A0A1W4W1Z1 A0A336LEQ5 A0A1W4WCL5 A0A0J9R6C9 Q7PLW4 A0A1B6KAW2 A0A0N8NZE4 A0A0Q9XHC3 A0A1A9W8U2 A0A0J9R7S8 Q5LJY0 A0A0J9R6Q2 A0A0J9TUA0 A0A1A9XBW7 A0A0J9R6L8 A0A1I8NQJ2 A0A1I8NQJ4 A0A088AGL0
A0A1Y9HFE4 A0A182Q635 A0A1S4F8Z8 Q17BT5 A0A182GKT4 A0A182VNA7 A0A182U315 A0A182MRU6 A0A2C9GR81 A0A182KKG1 A0A1L8DK38 W5JWA6 A0A182Y9M0 A0A182WUK8 A0A182J077 A0A182WE41 V5I898 A0A182FV54 A0A182N2P2 A0A1B0GMI3 A0A182PDA0 A0A084V9W0 A0A182K8I5 Q7PSJ7 A0A1B0GMI4 A0A2C9GR82 Q17BT4 A0A182JHY5 A0A1S4F8P6 D6WM87 A0A182Q6T2 W5JU26 A0A182UQI0 A0A2A3E1A4 A0A1W7R6T4 A0A088AGL8 A0A0T6AXF0 U4UKB9 A0A1I8MJH1 U4UTP4 A0A1B6D9C7 A0A0K8TR74 A0A034W6P0 A0A067R056 B0WBJ4 A0A067RBZ5 A0A1B6M4M0 A0A0K8US07 N6UFJ8 N6THY4 W8BQP8 A0A0K8W873 W8C8K6 A0A0Q9W6E5 A0A0L0BYT5 E2C4F0 A0A310S7P6 A0A0A9XWU9 A0A0A1XSB5 A0A224XMZ1 N6US54 A0A0M9A307 A0A0M3QZN1 A0A023EZI4 A0A2J7QAG0 A0A1W4W1Z0 A0A1W4W1Y6 A0A0L7RBA1 A0A1B6FZ48 A0A1W4WDX3 A0A1B6HL49 A0A021WWX0 A0A1B6JE47 B3MYI6 B4IM40 A0A1W4W1Z1 A0A336LEQ5 A0A1W4WCL5 A0A0J9R6C9 Q7PLW4 A0A1B6KAW2 A0A0N8NZE4 A0A0Q9XHC3 A0A1A9W8U2 A0A0J9R7S8 Q5LJY0 A0A0J9R6Q2 A0A0J9TUA0 A0A1A9XBW7 A0A0J9R6L8 A0A1I8NQJ2 A0A1I8NQJ4 A0A088AGL0
PDB
3V5U
E-value=1.95263e-05,
Score=116
Ontologies
GO
PANTHER
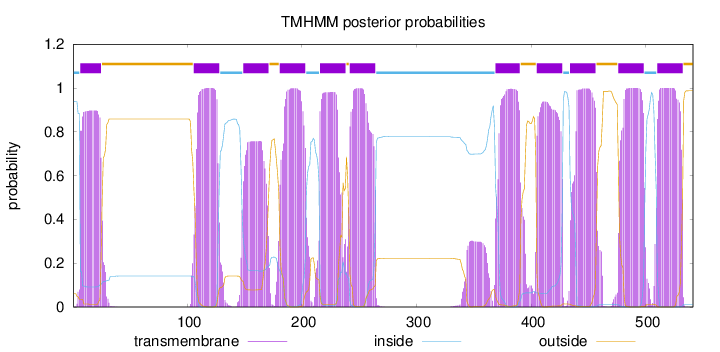
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.682174
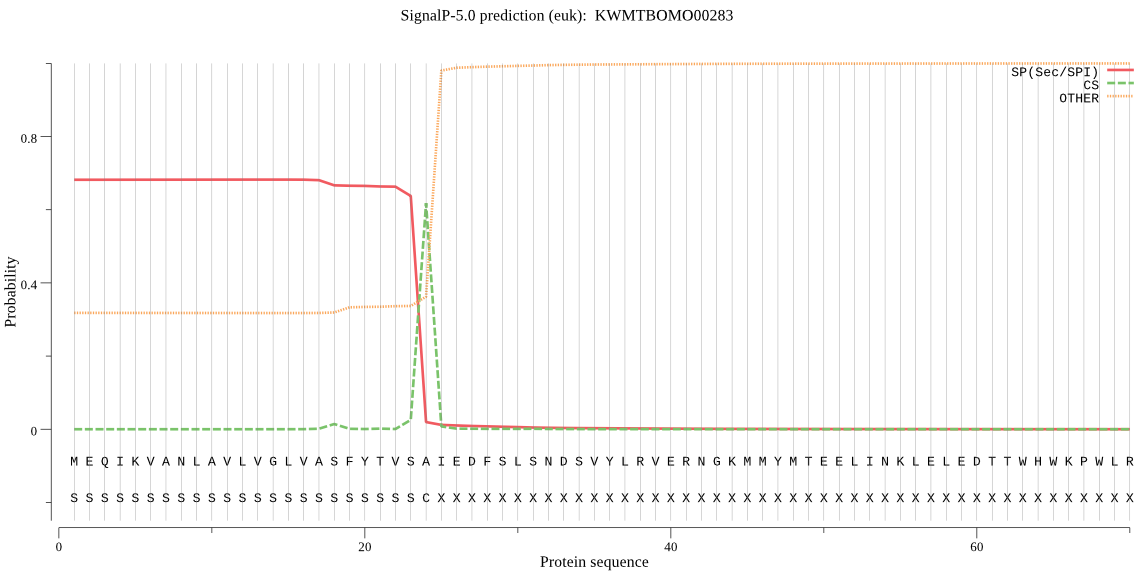
Length:
541
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
236.93128
Exp number, first 60 AAs:
17.77363
Total prob of N-in:
0.93900
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 25
outside
26 - 105
TMhelix
106 - 128
inside
129 - 148
TMhelix
149 - 171
outside
172 - 180
TMhelix
181 - 203
inside
204 - 215
TMhelix
216 - 238
outside
239 - 241
TMhelix
242 - 264
inside
265 - 368
TMhelix
369 - 390
outside
391 - 404
TMhelix
405 - 427
inside
428 - 433
TMhelix
434 - 456
outside
457 - 475
TMhelix
476 - 498
inside
499 - 509
TMhelix
510 - 532
outside
533 - 541
Population Genetic Test Statistics
Pi
6.417117
Theta
13.849783
Tajima's D
-0.641899
CLR
0.188851
CSRT
0.209589520523974
Interpretation
Uncertain
