Gene
KWMTBOMO00280
Pre Gene Modal
BGIBMGA000548
Annotation
PREDICTED:_RUS1_family_protein_C16orf58_homolog_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.028
Sequence
CDS
ATGTCTTCTCAACCAGAGGGTGAGATTATAGTCAAAGAAAAGTATGGATCTTCGAGTACCGAACGCATTTACATAAAACCACCCGAGCAAGCTATGGTTGTGGTGTATGATGAAACAAAGATCAATTACTTCAGGAACTGGTTCACGAATGTTTTTCTACCAAAAGGATATCCTGACAGTGTCAGCAGAGATTACTCAGCTTACCAGATATGGGATACAGCTCAAGCTTTCTGCAGCACCATAACAGGTACCTTAGCTACTCAAGAAGTGCTTAGAGGTGTTGGTGTTGGAGATACAACTGCTACTCCACTCGCTGCAACAGTGACATGGGTCATTAAAGACGGTTGTGGTCATCTAGGCAGGATACTCTTTGCTTTTACTCATGGAACGTGTTTGGATGCGTACAGCAAAAAATGGAGGCTTTACGCCGACATTTTAAATGACGCAGCGATGTGTATTGAAATTGCTCTTCCATACTTCAAAAACTACACTACAGTCGTACTGTGTGTGAGCACTGTTATGAAGGCAATTGTTGGAGTTTCAGGTGGTGCCACAAGAGCAGCTATGACGCAACACCATGCAGTAAGAGGCAACATGGCCGATGTTTCTGCCAAAGACTCAGCTCAAGAGACAGCTGTCAATCTTATTGCTTCTTTTACTGCTTTGTTTATTATCTCTGTTTTTGGGAGCAATGCGTTTATTTTCGTTTTCATAATGATGCTTCATATAGCGTTCAATTATTTTGCTGTAAGAGCTGTTTGTTTACGGACTATGAATGAACCTAGATTTTTGGAGTTTATCAACATATATTTAACGAAAGAAAGAATTGCAACACCGTGTGAAGTGAACAGACGCGAACCAGTCGCTTTTTATCAGATGGGCTCGCATACATTAGACCTCAAATTGTGTGGTTTCCAAATTAAAATGGGAGTGTCCATAAAGGATCTCATTAAAAAAAAATATCCAAAGGCGTCACACTTAAAGAACATCAAAGACGTCTACGAGAACCGAAATTATATCATAATCCCCAATGTTCAAGACCGGAAAATGTATGTGCTGCTTAATGAAAAGGCAATGAACGATGATATTCTGCGTGCTTACTTTCATGCTGTTCTCCTTTCCGTTATCATATGTGTAATCAACGATGTGCCCTTGCCCGTTTACCAAGCTCCGAATGAGGAGAGACCTTTCGCGCAGGTATGCCGCACAATACAAACAACTGATTGGAGCCGGGAACCTATAGAGACTCACGTTTTTAATGAATTCAAATTTGAGCCTTCTTACGACCTAATGAAATACGTTAATAGAATTGTTCAACGAGAATGGATGAGAATTAAAACGGGTATGATGCGCATAGGTTGGGATCTCAATAAACACCTGCTGATGGTAGACGAATGGCGTATTTCTTCGGTAAAACCACTTTCTCAATACGCGCCAATAGAGGGTAGAGTATCAACAAAGGACTATGTGATCAGTGAGGCGGATGCAAAACTTACGAAAGAATTGCTTATGGAATTAGAACATGGTGATGGGGTCACTTACGGCGTAGTAGAGACCAAGCCTTTGATGCATTACAAGAAAGAAGCTTCCAAACGACGAGTCCACGATATCGGCACTGAAACAGAATCACCAGAAATCGAAGAATATCGTGAACCGAATACAGAAGCGGAAGTTAAGGCTGAAGTACCGAAAGAAAACAGGAAAAAAGACTGA
Protein
MSSQPEGEIIVKEKYGSSSTERIYIKPPEQAMVVVYDETKINYFRNWFTNVFLPKGYPDSVSRDYSAYQIWDTAQAFCSTITGTLATQEVLRGVGVGDTTATPLAATVTWVIKDGCGHLGRILFAFTHGTCLDAYSKKWRLYADILNDAAMCIEIALPYFKNYTTVVLCVSTVMKAIVGVSGGATRAAMTQHHAVRGNMADVSAKDSAQETAVNLIASFTALFIISVFGSNAFIFVFIMMLHIAFNYFAVRAVCLRTMNEPRFLEFINIYLTKERIATPCEVNRREPVAFYQMGSHTLDLKLCGFQIKMGVSIKDLIKKKYPKASHLKNIKDVYENRNYIIIPNVQDRKMYVLLNEKAMNDDILRAYFHAVLLSVIICVINDVPLPVYQAPNEERPFAQVCRTIQTTDWSREPIETHVFNEFKFEPSYDLMKYVNRIVQREWMRIKTGMMRIGWDLNKHLLMVDEWRISSVKPLSQYAPIEGRVSTKDYVISEADAKLTKELLMELEHGDGVTYGVVETKPLMHYKKEASKRRVHDIGTETESPEIEEYREPNTEAEVKAEVPKENRKKD
Summary
Uniprot
A0A1E1WDA0
A0A2A4JR68
A0A2H1VNW5
A0A212ETE1
A0A0L7LKR8
A0A1B6IFU3
+ More
A0A2J7PJZ6 A0A067R4N1 A0A1B6EI35 A0A154PCS9 A0A088A6C4 A0A195C320 A0A195EYP2 A0A195DIN5 E9J1X9 A0A151X3X6 E2BJZ5 F4W783 A0A1W4VMW9 A0A158NJ45 A0A026WBJ1 A0A1Y1NEA1 A0A195BJU2 A0A023F0A2 Q29LS8 B4G7G5 B4LSK5 B4KJQ7 B4P9W8 W8C9B3 B4Q8X1 D6X0W0 A0A3B0K7C2 Q9VJ63 A0A0A1WEY8 B3NLB0 B4I5J5 B3MM45 A0A0P4VME4 B4JBZ0 A0A2A3EBP0 E1ZVM4 U4UHM2 V5GPM4 A0A0L7RJL4 N6TCD4 A0A1I8MJ64 A0A224XJ25 A0A162QBV2 A0A0V0G5D6 A0A1B0GAU7 A0A0L0CFV1 A0A1A9VI68 A0A0P4XMP8 A0A1A9ZEV7 B4MZN5 A0A1A9YR54 A0A1B0BDR4 A0A1A9WN23 A0A1S3J6L0 A0A336M0X6 A0A182NNZ4 A0A0N8DVC1
A0A2J7PJZ6 A0A067R4N1 A0A1B6EI35 A0A154PCS9 A0A088A6C4 A0A195C320 A0A195EYP2 A0A195DIN5 E9J1X9 A0A151X3X6 E2BJZ5 F4W783 A0A1W4VMW9 A0A158NJ45 A0A026WBJ1 A0A1Y1NEA1 A0A195BJU2 A0A023F0A2 Q29LS8 B4G7G5 B4LSK5 B4KJQ7 B4P9W8 W8C9B3 B4Q8X1 D6X0W0 A0A3B0K7C2 Q9VJ63 A0A0A1WEY8 B3NLB0 B4I5J5 B3MM45 A0A0P4VME4 B4JBZ0 A0A2A3EBP0 E1ZVM4 U4UHM2 V5GPM4 A0A0L7RJL4 N6TCD4 A0A1I8MJ64 A0A224XJ25 A0A162QBV2 A0A0V0G5D6 A0A1B0GAU7 A0A0L0CFV1 A0A1A9VI68 A0A0P4XMP8 A0A1A9ZEV7 B4MZN5 A0A1A9YR54 A0A1B0BDR4 A0A1A9WN23 A0A1S3J6L0 A0A336M0X6 A0A182NNZ4 A0A0N8DVC1
Pubmed
22118469
26227816
24845553
21282665
20798317
21719571
+ More
21347285 24508170 30249741 28004739 25474469 15632085 17994087 17550304 24495485 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 27129103 23537049 25315136 26108605
21347285 24508170 30249741 28004739 25474469 15632085 17994087 17550304 24495485 22936249 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 27129103 23537049 25315136 26108605
EMBL
GDQN01006090
JAT84964.1
NWSH01000732
PCG74527.1
ODYU01003579
SOQ42520.1
+ More
AGBW02012593 OWR44763.1 JTDY01000766 KOB75959.1 GECU01021923 JAS85783.1 NEVH01024944 PNF16648.1 KK852704 KDR18057.1 GECZ01032209 JAS37560.1 KQ434874 KZC09706.1 KQ978317 KYM95259.1 KQ981920 KYN33027.1 KQ980804 KYN12758.1 GL767674 EFZ13253.1 KQ982559 KYQ54979.1 GL448708 EFN84004.1 GL887813 EGI69967.1 ADTU01017450 KK107293 QOIP01000005 EZA53425.1 RLU22240.1 GEZM01007427 JAV95210.1 KQ976453 KYM85456.1 GBBI01003782 JAC14930.1 CH379060 EAL33967.1 CH479180 EDW29298.1 CH940649 EDW64827.1 CH933807 EDW11502.1 CM000158 EDW90309.1 GAMC01002310 GAMC01002309 JAC04247.1 CM000361 CM002910 EDX05403.1 KMY90861.1 KQ971372 EFA09542.2 OUUW01000004 SPP79408.1 AE014134 BT133447 AAF53692.2 AFH89820.1 AHN54570.1 GBXI01016835 JAC97456.1 CH954179 EDV54760.1 CH480822 EDW55651.1 CH902620 EDV30860.1 GDKW01000282 JAI56313.1 CH916368 EDW04093.1 KZ288300 PBC28884.1 GL434548 EFN74793.1 KB632328 ERL92532.1 GALX01002397 JAB66069.1 KQ414582 KOC70916.1 APGK01043270 KB741014 ENN75403.1 GFTR01006608 JAW09818.1 LRGB01000359 KZS19548.1 GECL01002918 JAP03206.1 CCAG010020051 JRES01000438 KNC31288.1 GDIP01241811 JAI81590.1 CH963920 EDW77820.1 JXJN01012634 UFQT01000272 SSX22649.1 GDIQ01088300 JAN06437.1
AGBW02012593 OWR44763.1 JTDY01000766 KOB75959.1 GECU01021923 JAS85783.1 NEVH01024944 PNF16648.1 KK852704 KDR18057.1 GECZ01032209 JAS37560.1 KQ434874 KZC09706.1 KQ978317 KYM95259.1 KQ981920 KYN33027.1 KQ980804 KYN12758.1 GL767674 EFZ13253.1 KQ982559 KYQ54979.1 GL448708 EFN84004.1 GL887813 EGI69967.1 ADTU01017450 KK107293 QOIP01000005 EZA53425.1 RLU22240.1 GEZM01007427 JAV95210.1 KQ976453 KYM85456.1 GBBI01003782 JAC14930.1 CH379060 EAL33967.1 CH479180 EDW29298.1 CH940649 EDW64827.1 CH933807 EDW11502.1 CM000158 EDW90309.1 GAMC01002310 GAMC01002309 JAC04247.1 CM000361 CM002910 EDX05403.1 KMY90861.1 KQ971372 EFA09542.2 OUUW01000004 SPP79408.1 AE014134 BT133447 AAF53692.2 AFH89820.1 AHN54570.1 GBXI01016835 JAC97456.1 CH954179 EDV54760.1 CH480822 EDW55651.1 CH902620 EDV30860.1 GDKW01000282 JAI56313.1 CH916368 EDW04093.1 KZ288300 PBC28884.1 GL434548 EFN74793.1 KB632328 ERL92532.1 GALX01002397 JAB66069.1 KQ414582 KOC70916.1 APGK01043270 KB741014 ENN75403.1 GFTR01006608 JAW09818.1 LRGB01000359 KZS19548.1 GECL01002918 JAP03206.1 CCAG010020051 JRES01000438 KNC31288.1 GDIP01241811 JAI81590.1 CH963920 EDW77820.1 JXJN01012634 UFQT01000272 SSX22649.1 GDIQ01088300 JAN06437.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000235965
UP000027135
UP000076502
+ More
UP000005203 UP000078542 UP000078541 UP000078492 UP000075809 UP000008237 UP000007755 UP000192221 UP000005205 UP000053097 UP000279307 UP000078540 UP000001819 UP000008744 UP000008792 UP000009192 UP000002282 UP000000304 UP000007266 UP000268350 UP000000803 UP000008711 UP000001292 UP000007801 UP000001070 UP000242457 UP000000311 UP000030742 UP000053825 UP000019118 UP000095301 UP000076858 UP000092444 UP000037069 UP000078200 UP000092445 UP000007798 UP000092443 UP000092460 UP000091820 UP000085678 UP000075884
UP000005203 UP000078542 UP000078541 UP000078492 UP000075809 UP000008237 UP000007755 UP000192221 UP000005205 UP000053097 UP000279307 UP000078540 UP000001819 UP000008744 UP000008792 UP000009192 UP000002282 UP000000304 UP000007266 UP000268350 UP000000803 UP000008711 UP000001292 UP000007801 UP000001070 UP000242457 UP000000311 UP000030742 UP000053825 UP000019118 UP000095301 UP000076858 UP000092444 UP000037069 UP000078200 UP000092445 UP000007798 UP000092443 UP000092460 UP000091820 UP000085678 UP000075884
PRIDE
SUPFAM
SSF158573
SSF158573
ProteinModelPortal
A0A1E1WDA0
A0A2A4JR68
A0A2H1VNW5
A0A212ETE1
A0A0L7LKR8
A0A1B6IFU3
+ More
A0A2J7PJZ6 A0A067R4N1 A0A1B6EI35 A0A154PCS9 A0A088A6C4 A0A195C320 A0A195EYP2 A0A195DIN5 E9J1X9 A0A151X3X6 E2BJZ5 F4W783 A0A1W4VMW9 A0A158NJ45 A0A026WBJ1 A0A1Y1NEA1 A0A195BJU2 A0A023F0A2 Q29LS8 B4G7G5 B4LSK5 B4KJQ7 B4P9W8 W8C9B3 B4Q8X1 D6X0W0 A0A3B0K7C2 Q9VJ63 A0A0A1WEY8 B3NLB0 B4I5J5 B3MM45 A0A0P4VME4 B4JBZ0 A0A2A3EBP0 E1ZVM4 U4UHM2 V5GPM4 A0A0L7RJL4 N6TCD4 A0A1I8MJ64 A0A224XJ25 A0A162QBV2 A0A0V0G5D6 A0A1B0GAU7 A0A0L0CFV1 A0A1A9VI68 A0A0P4XMP8 A0A1A9ZEV7 B4MZN5 A0A1A9YR54 A0A1B0BDR4 A0A1A9WN23 A0A1S3J6L0 A0A336M0X6 A0A182NNZ4 A0A0N8DVC1
A0A2J7PJZ6 A0A067R4N1 A0A1B6EI35 A0A154PCS9 A0A088A6C4 A0A195C320 A0A195EYP2 A0A195DIN5 E9J1X9 A0A151X3X6 E2BJZ5 F4W783 A0A1W4VMW9 A0A158NJ45 A0A026WBJ1 A0A1Y1NEA1 A0A195BJU2 A0A023F0A2 Q29LS8 B4G7G5 B4LSK5 B4KJQ7 B4P9W8 W8C9B3 B4Q8X1 D6X0W0 A0A3B0K7C2 Q9VJ63 A0A0A1WEY8 B3NLB0 B4I5J5 B3MM45 A0A0P4VME4 B4JBZ0 A0A2A3EBP0 E1ZVM4 U4UHM2 V5GPM4 A0A0L7RJL4 N6TCD4 A0A1I8MJ64 A0A224XJ25 A0A162QBV2 A0A0V0G5D6 A0A1B0GAU7 A0A0L0CFV1 A0A1A9VI68 A0A0P4XMP8 A0A1A9ZEV7 B4MZN5 A0A1A9YR54 A0A1B0BDR4 A0A1A9WN23 A0A1S3J6L0 A0A336M0X6 A0A182NNZ4 A0A0N8DVC1
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
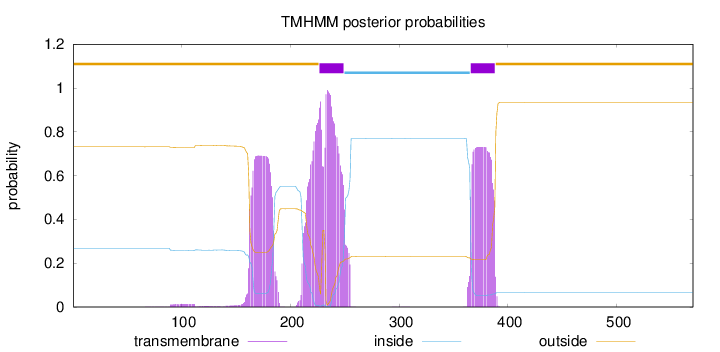
Length:
570
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
61.92428
Exp number, first 60 AAs:
0.00059
Total prob of N-in:
0.26834
outside
1 - 226
TMhelix
227 - 249
inside
250 - 365
TMhelix
366 - 388
outside
389 - 570
Population Genetic Test Statistics
Pi
5.61681
Theta
5.030776
Tajima's D
-1.374387
CLR
0.094857
CSRT
0.0765961701914904
Interpretation
Uncertain
