Gene
KWMTBOMO00279 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000672
Annotation
putative_citrate_synthase_[Danaus_plexippus]
Full name
Citrate synthase
+ More
Probable citrate synthase 2, mitochondrial
Probable citrate synthase 1, mitochondrial
Probable citrate synthase, mitochondrial
Probable citrate synthase 2, mitochondrial
Probable citrate synthase 1, mitochondrial
Probable citrate synthase, mitochondrial
Alternative Name
Protein knockdown
Location in the cell
Mitochondrial Reliability : 3.614
Sequence
CDS
ATGGCTCTATTCAGGATCACATCTTCAAGACTCGTAGAATTGCAGAAAGCATGTCCGACTGCGACAGTTCTACTTCGAGGTCTAAGCGCTGAACAGACGAATTTAAAAAGTATTCTCCAGGAAAAGATTCCCAAGGAACAAGAGAAGATCCGCGAATTCCGAAAGAAGCATGGTTCCACTAAAGTCGGAGAGGTCACAGTTGATATGATGTACGGTGGCATGCGTGGAATCAAAGGTCTGGTTTGGGAAACCTCTGTGCTGGATGCCGATGAAGGAATCCGTTTCCGTGGTCTATCCATCCCTGAGTGCCAACAGCAACTGCCCAAGGCTAAGGGTGGAGAAGAACCACTACCCGAAGGTCTTTTCTGGCTTCTCGTCACTGGCGATATACCCACCGAAGCGCAAGCTAAAGCGCTGTCTAAAGAATGGGCAGCGAGGGCGGAGCTACCGGCTCACGTAGTGACAATGTTGAACAATATGCCCGGCAAACTGCATCCCATGTCACAGTTCTCGGCTGCCGTCACCGCACTCAACAGTGAATCTAAATTCGCTAAAGCCTACTCAGAGGGCGTGCACAAATCCAAGTATTGGGAGTACGTGTACGAAGACACAATGAACCTTATCGCGAAACTGCCCGTGATCGCCGCCACTATCTATCGCAACGTGTACCGCGATGGCAAGGGCATCGGCGCCATCGACGACAACAAGGACTGGTCCGCGAACTACTGCAACATGCTCGGCTTCGACGACCCGCAGTTCACCGAACTCATGCGTCTCTACCTCACCATACACAGTGACCACGAGGGTGGCAACGTTTCTGCGCACACTACGCATTTAGTGGGTTCTGCTCTTAGTGATCCTTACTTATCTTTCGCCGCTGGACTCAATGGTCTTGCTGGACCACTTCACGGCCTGGCTAATCAAGAGGTTCTGGTATGGCTCGAGAAACTGCGCAAACAAGTTGGTGATAATTTCACAGAAGAGCAACTCAAAGAATTCATCTGGAAAACACTTAAATCTGGTCAGGTTGTACCTGGTTACGGTCACGCAGTACTTAGAAAAACTGATCCAAGATATACTTGCCAGCGTGAGTTTGCTCTTAAGCACTTACCCAATGACCCATTATTCAAGCTGGTCGCTGCTGTTTACAAGGTTGTTCCGCCGATCCTCACCGAACTTGGCAAAGTCAAGAACCCATGGCCTAATGTAGACTCCCATTCGGGAGTTCTTTTGCAGTATTATGGTCTGAAGGAGATGAACTACTATACAGTAATGTTTGGAGTGTCCCGAGCGTTGGGTGTTCTCGCTCAGTTGATTTGGTCCCGTGCGCTCGGACTTCCCATTGAGAGACCAAAGTCGCTCAGCACGGAGATGCTTATGAAACAAGTCGGGAAGTAA
Protein
MALFRITSSRLVELQKACPTATVLLRGLSAEQTNLKSILQEKIPKEQEKIREFRKKHGSTKVGEVTVDMMYGGMRGIKGLVWETSVLDADEGIRFRGLSIPECQQQLPKAKGGEEPLPEGLFWLLVTGDIPTEAQAKALSKEWAARAELPAHVVTMLNNMPGKLHPMSQFSAAVTALNSESKFAKAYSEGVHKSKYWEYVYEDTMNLIAKLPVIAATIYRNVYRDGKGIGAIDDNKDWSANYCNMLGFDDPQFTELMRLYLTIHSDHEGGNVSAHTTHLVGSALSDPYLSFAAGLNGLAGPLHGLANQEVLVWLEKLRKQVGDNFTEEQLKEFIWKTLKSGQVVPGYGHAVLRKTDPRYTCQREFALKHLPNDPLFKLVAAVYKVVPPILTELGKVKNPWPNVDSHSGVLLQYYGLKEMNYYTVMFGVSRALGVLAQLIWSRALGLPIERPKSLSTEMLMKQVGK
Summary
Description
Plays a role in controlling neuronal activity and seizure susceptibility.
Catalytic Activity
acetyl-CoA + H2O + oxaloacetate = citrate + CoA + H(+)
Subunit
Homodimer.
Miscellaneous
Citrate synthase is found in nearly all cells capable of oxidative metabolism.
Mutation of kdn causes a behavioral mutation ('bang sensitivity' = temporarily paralysis in response to a physical jolt). Flies lacking kdn exhibit alterations in neuronal firing patterns in the giant fiber (GF) pathway.
Mutation of kdn causes a behavioral mutation ('bang sensitivity' = temporarily paralysis in response to a physical jolt). Flies lacking kdn exhibit alterations in neuronal firing patterns in the giant fiber (GF) pathway.
Similarity
Belongs to the citrate synthase family.
Keywords
Complete proteome
Mitochondrion
Reference proteome
Transferase
Transit peptide
Tricarboxylic acid cycle
Alternative splicing
Behavior
Feature
chain Probable citrate synthase 2, mitochondrial
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9ITU2
A0A2A4JPM9
A0A212ETD4
A0A2A4JNS8
A0A2H1VNY9
A0A194QWB0
+ More
A0A194PS44 J3JV43 U4UCM5 A0A1Y1K9C2 A0A1W4WQY7 D6WMM4 A0A023ESP1 N6U883 A0A2J7RJY2 A0A084WJ30 A0A2Z5RE60 Q16P20 U5EUY0 A0A182PV63 Q17GM7 V5G175 A0A1I8JTB6 A0A182UVT4 A0A182KTE7 Q7PV48 A0A182RSS8 A0A336M4V0 A0A182N3L6 A0A182Q2D7 A0A1Y9IVZ9 A0A1Y9J0F6 T1E9V8 A0A2M3Z0V5 T1E2L3 A0A2M4A7U9 A0A2M4A7S6 A0A2M4CUN3 A0A1Q3FUN5 A0A1W5YLL6 A0A182K1V2 A0A182FVT2 A0A182GKM3 A0A182YCD2 A0A1J1IJG4 A0A3Q0IKK7 A0A182M257 A0A1Q3FV06 A0A182JN40 W5J9I7 A0A1W6EVT2 A0A3L8D9H6 A0A1B6LTM4 A0A1B6ERI7 A0A1S3CUV0 A0A1B6FW63 A0A0K8TSU1 A0A1L8E0W5 A0A1L8E196 A0A158NK90 A0A1L8E0T5 A0A1L8EE92 A0A067RFE0 A0A1L8EEN0 T1PDY8 B4N2K9 A0A069DTW1 A0A0P4VSB3 R4G5K7 A0A1B0D970 A0A0V0G5W9 A0A1B6CDS0 A0A224XBZ6 A0A0A1XDV0 A0A0J7NLK9 V5SJ12 A0A195FBS5 D3TQS9 A0A0P4WHU5 Q0QHL3 A0A1I8PUP1 W8CC68 A0A0A9VZC2 A0A023F8N9 R4WJI6 E2BRV0 A0A026X2M5 A0A0P8XS56 A0A151X6U5 A0A1W4VF19 A0A1I9WLM9 E2AVF4 C6TP50 B3NXS1 A0A0M5JAP7 A0A1B6JC96 A0A0R3NYW2 E0VRF1 Q9W401 B4L2G7
A0A194PS44 J3JV43 U4UCM5 A0A1Y1K9C2 A0A1W4WQY7 D6WMM4 A0A023ESP1 N6U883 A0A2J7RJY2 A0A084WJ30 A0A2Z5RE60 Q16P20 U5EUY0 A0A182PV63 Q17GM7 V5G175 A0A1I8JTB6 A0A182UVT4 A0A182KTE7 Q7PV48 A0A182RSS8 A0A336M4V0 A0A182N3L6 A0A182Q2D7 A0A1Y9IVZ9 A0A1Y9J0F6 T1E9V8 A0A2M3Z0V5 T1E2L3 A0A2M4A7U9 A0A2M4A7S6 A0A2M4CUN3 A0A1Q3FUN5 A0A1W5YLL6 A0A182K1V2 A0A182FVT2 A0A182GKM3 A0A182YCD2 A0A1J1IJG4 A0A3Q0IKK7 A0A182M257 A0A1Q3FV06 A0A182JN40 W5J9I7 A0A1W6EVT2 A0A3L8D9H6 A0A1B6LTM4 A0A1B6ERI7 A0A1S3CUV0 A0A1B6FW63 A0A0K8TSU1 A0A1L8E0W5 A0A1L8E196 A0A158NK90 A0A1L8E0T5 A0A1L8EE92 A0A067RFE0 A0A1L8EEN0 T1PDY8 B4N2K9 A0A069DTW1 A0A0P4VSB3 R4G5K7 A0A1B0D970 A0A0V0G5W9 A0A1B6CDS0 A0A224XBZ6 A0A0A1XDV0 A0A0J7NLK9 V5SJ12 A0A195FBS5 D3TQS9 A0A0P4WHU5 Q0QHL3 A0A1I8PUP1 W8CC68 A0A0A9VZC2 A0A023F8N9 R4WJI6 E2BRV0 A0A026X2M5 A0A0P8XS56 A0A151X6U5 A0A1W4VF19 A0A1I9WLM9 E2AVF4 C6TP50 B3NXS1 A0A0M5JAP7 A0A1B6JC96 A0A0R3NYW2 E0VRF1 Q9W401 B4L2G7
EC Number
2.3.3.16
Pubmed
19121390
22118469
26354079
22516182
23537049
28004739
+ More
18362917 19820115 24945155 24438588 26760975 17510324 20966253 12364791 14747013 17210077 24330624 26483478 25244985 20920257 23761445 30249741 26369729 21347285 24845553 25315136 17994087 18057021 26334808 27129103 25830018 20353571 16907828 24495485 25401762 26823975 25474469 23691247 20798317 24508170 27538518 15632085 23185243 20566863 10731132 12537572 12537569 14680800 16648587
18362917 19820115 24945155 24438588 26760975 17510324 20966253 12364791 14747013 17210077 24330624 26483478 25244985 20920257 23761445 30249741 26369729 21347285 24845553 25315136 17994087 18057021 26334808 27129103 25830018 20353571 16907828 24495485 25401762 26823975 25474469 23691247 20798317 24508170 27538518 15632085 23185243 20566863 10731132 12537572 12537569 14680800 16648587
EMBL
BABH01006978
NWSH01000884
PCG73716.1
AGBW02012593
OWR44762.1
PCG73715.1
+ More
ODYU01003579 SOQ42518.1 KQ461108 KPJ09250.1 KQ459594 KPI96132.1 BT127109 AEE62071.1 KB632297 ERL91704.1 GEZM01088604 JAV58072.1 KQ971343 EFA03279.1 GAPW01001305 JAC12293.1 APGK01044816 APGK01044817 APGK01044818 KB741028 ENN74802.1 NEVH01002982 PNF41146.1 ATLV01023968 KE525347 KFB50224.1 FX983734 BAX07233.1 CH477799 GANO01001257 JAB58614.1 CH477258 GALX01004701 JAB63765.1 APCN01002450 AAAB01008986 EAA00454.5 UFQS01000559 UFQT01000559 SSX04940.1 SSX25302.1 AXCN02001106 GAMD01001563 JAB00028.1 GGFM01001411 MBW22162.1 GALA01001160 JAA93692.1 GGFK01003499 MBW36820.1 GGFK01003508 MBW36829.1 GGFL01004370 MBW68548.1 GFDL01003726 JAV31319.1 KX147674 ARI45078.1 JXUM01070227 JXUM01070228 JXUM01070229 JXUM01070230 JXUM01070231 JXUM01070232 KQ562595 KXJ75524.1 CVRI01000054 CRL00383.1 AXCM01005459 GFDL01003729 JAV31316.1 ADMH02001988 ETN60073.1 KY563412 ARK19821.1 QOIP01000011 RLU16539.1 GEBQ01012937 JAT27040.1 GECZ01029227 JAS40542.1 GECZ01015486 JAS54283.1 GDAI01000164 JAI17439.1 GFDF01001743 JAV12341.1 GFDF01001742 JAV12342.1 ADTU01018740 ADTU01018741 GFDF01001744 JAV12340.1 GFDG01001880 JAV16919.1 KK852498 KDR22581.1 GFDG01001695 JAV17104.1 KA646158 AFP60787.1 CH963925 EDW78598.1 KRF98915.1 GBGD01001356 JAC87533.1 GDKW01003700 JAI52895.1 ACPB03000853 GAHY01000455 JAA77055.1 AJVK01004260 AJVK01004261 GECL01002680 JAP03444.1 GEDC01025705 JAS11593.1 GFTR01006501 JAW09925.1 GBXI01012403 GBXI01004713 JAD01889.1 JAD09579.1 LBMM01003579 KMQ93360.1 KF647629 AHB50491.1 KQ981693 KYN37662.1 EZ423781 ADD20057.1 GDRN01071936 JAI63621.1 DQ374006 GAMC01004166 GAMC01004165 JAC02390.1 GBHO01045634 GBRD01011409 GDHC01020673 JAF97969.1 JAG54415.1 JAP97955.1 GBBI01000972 JAC17740.1 AK417710 BAN20925.1 GL450042 EFN81583.1 KK107021 EZA62343.1 CH902630 KPU77480.1 KQ982472 KYQ56115.1 KU932413 APA34049.1 GL443090 EFN62588.1 BT099536 ACU32620.1 CH954180 EDV47372.1 CP012528 ALC49224.1 GECU01010846 JAS96860.1 CH379063 KRT06226.1 DS235465 EEB15957.1 AE014298 AY089318 CH933810 EDW06843.1
ODYU01003579 SOQ42518.1 KQ461108 KPJ09250.1 KQ459594 KPI96132.1 BT127109 AEE62071.1 KB632297 ERL91704.1 GEZM01088604 JAV58072.1 KQ971343 EFA03279.1 GAPW01001305 JAC12293.1 APGK01044816 APGK01044817 APGK01044818 KB741028 ENN74802.1 NEVH01002982 PNF41146.1 ATLV01023968 KE525347 KFB50224.1 FX983734 BAX07233.1 CH477799 GANO01001257 JAB58614.1 CH477258 GALX01004701 JAB63765.1 APCN01002450 AAAB01008986 EAA00454.5 UFQS01000559 UFQT01000559 SSX04940.1 SSX25302.1 AXCN02001106 GAMD01001563 JAB00028.1 GGFM01001411 MBW22162.1 GALA01001160 JAA93692.1 GGFK01003499 MBW36820.1 GGFK01003508 MBW36829.1 GGFL01004370 MBW68548.1 GFDL01003726 JAV31319.1 KX147674 ARI45078.1 JXUM01070227 JXUM01070228 JXUM01070229 JXUM01070230 JXUM01070231 JXUM01070232 KQ562595 KXJ75524.1 CVRI01000054 CRL00383.1 AXCM01005459 GFDL01003729 JAV31316.1 ADMH02001988 ETN60073.1 KY563412 ARK19821.1 QOIP01000011 RLU16539.1 GEBQ01012937 JAT27040.1 GECZ01029227 JAS40542.1 GECZ01015486 JAS54283.1 GDAI01000164 JAI17439.1 GFDF01001743 JAV12341.1 GFDF01001742 JAV12342.1 ADTU01018740 ADTU01018741 GFDF01001744 JAV12340.1 GFDG01001880 JAV16919.1 KK852498 KDR22581.1 GFDG01001695 JAV17104.1 KA646158 AFP60787.1 CH963925 EDW78598.1 KRF98915.1 GBGD01001356 JAC87533.1 GDKW01003700 JAI52895.1 ACPB03000853 GAHY01000455 JAA77055.1 AJVK01004260 AJVK01004261 GECL01002680 JAP03444.1 GEDC01025705 JAS11593.1 GFTR01006501 JAW09925.1 GBXI01012403 GBXI01004713 JAD01889.1 JAD09579.1 LBMM01003579 KMQ93360.1 KF647629 AHB50491.1 KQ981693 KYN37662.1 EZ423781 ADD20057.1 GDRN01071936 JAI63621.1 DQ374006 GAMC01004166 GAMC01004165 JAC02390.1 GBHO01045634 GBRD01011409 GDHC01020673 JAF97969.1 JAG54415.1 JAP97955.1 GBBI01000972 JAC17740.1 AK417710 BAN20925.1 GL450042 EFN81583.1 KK107021 EZA62343.1 CH902630 KPU77480.1 KQ982472 KYQ56115.1 KU932413 APA34049.1 GL443090 EFN62588.1 BT099536 ACU32620.1 CH954180 EDV47372.1 CP012528 ALC49224.1 GECU01010846 JAS96860.1 CH379063 KRT06226.1 DS235465 EEB15957.1 AE014298 AY089318 CH933810 EDW06843.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000030742
+ More
UP000192223 UP000007266 UP000019118 UP000235965 UP000030765 UP000008820 UP000075885 UP000075840 UP000075903 UP000075882 UP000007062 UP000075900 UP000075884 UP000075886 UP000075920 UP000076407 UP000075881 UP000069272 UP000069940 UP000249989 UP000076408 UP000183832 UP000079169 UP000075883 UP000075880 UP000000673 UP000279307 UP000005205 UP000027135 UP000095301 UP000007798 UP000015103 UP000092462 UP000036403 UP000078541 UP000092444 UP000095300 UP000008237 UP000053097 UP000007801 UP000075809 UP000192221 UP000000311 UP000008711 UP000092553 UP000001819 UP000009046 UP000000803 UP000009192
UP000192223 UP000007266 UP000019118 UP000235965 UP000030765 UP000008820 UP000075885 UP000075840 UP000075903 UP000075882 UP000007062 UP000075900 UP000075884 UP000075886 UP000075920 UP000076407 UP000075881 UP000069272 UP000069940 UP000249989 UP000076408 UP000183832 UP000079169 UP000075883 UP000075880 UP000000673 UP000279307 UP000005205 UP000027135 UP000095301 UP000007798 UP000015103 UP000092462 UP000036403 UP000078541 UP000092444 UP000095300 UP000008237 UP000053097 UP000007801 UP000075809 UP000192221 UP000000311 UP000008711 UP000092553 UP000001819 UP000009046 UP000000803 UP000009192
Interpro
IPR019810
Citrate_synthase_AS
+ More
IPR016143 Citrate_synth-like_sm_a-sub
IPR036969 Citrate_synthase_sf
IPR010109 Citrate_synthase_euk
IPR002020 Citrate_synthase
IPR016142 Citrate_synth-like_lrg_a-sub
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR035518 DPG_synthase
IPR001173 Glyco_trans_2-like
IPR029044 Nucleotide-diphossugar_trans
IPR003169 GYF
IPR035445 GYF-like_dom_sf
IPR016143 Citrate_synth-like_sm_a-sub
IPR036969 Citrate_synthase_sf
IPR010109 Citrate_synthase_euk
IPR002020 Citrate_synthase
IPR016142 Citrate_synth-like_lrg_a-sub
IPR011989 ARM-like
IPR016024 ARM-type_fold
IPR035518 DPG_synthase
IPR001173 Glyco_trans_2-like
IPR029044 Nucleotide-diphossugar_trans
IPR003169 GYF
IPR035445 GYF-like_dom_sf
ProteinModelPortal
H9ITU2
A0A2A4JPM9
A0A212ETD4
A0A2A4JNS8
A0A2H1VNY9
A0A194QWB0
+ More
A0A194PS44 J3JV43 U4UCM5 A0A1Y1K9C2 A0A1W4WQY7 D6WMM4 A0A023ESP1 N6U883 A0A2J7RJY2 A0A084WJ30 A0A2Z5RE60 Q16P20 U5EUY0 A0A182PV63 Q17GM7 V5G175 A0A1I8JTB6 A0A182UVT4 A0A182KTE7 Q7PV48 A0A182RSS8 A0A336M4V0 A0A182N3L6 A0A182Q2D7 A0A1Y9IVZ9 A0A1Y9J0F6 T1E9V8 A0A2M3Z0V5 T1E2L3 A0A2M4A7U9 A0A2M4A7S6 A0A2M4CUN3 A0A1Q3FUN5 A0A1W5YLL6 A0A182K1V2 A0A182FVT2 A0A182GKM3 A0A182YCD2 A0A1J1IJG4 A0A3Q0IKK7 A0A182M257 A0A1Q3FV06 A0A182JN40 W5J9I7 A0A1W6EVT2 A0A3L8D9H6 A0A1B6LTM4 A0A1B6ERI7 A0A1S3CUV0 A0A1B6FW63 A0A0K8TSU1 A0A1L8E0W5 A0A1L8E196 A0A158NK90 A0A1L8E0T5 A0A1L8EE92 A0A067RFE0 A0A1L8EEN0 T1PDY8 B4N2K9 A0A069DTW1 A0A0P4VSB3 R4G5K7 A0A1B0D970 A0A0V0G5W9 A0A1B6CDS0 A0A224XBZ6 A0A0A1XDV0 A0A0J7NLK9 V5SJ12 A0A195FBS5 D3TQS9 A0A0P4WHU5 Q0QHL3 A0A1I8PUP1 W8CC68 A0A0A9VZC2 A0A023F8N9 R4WJI6 E2BRV0 A0A026X2M5 A0A0P8XS56 A0A151X6U5 A0A1W4VF19 A0A1I9WLM9 E2AVF4 C6TP50 B3NXS1 A0A0M5JAP7 A0A1B6JC96 A0A0R3NYW2 E0VRF1 Q9W401 B4L2G7
A0A194PS44 J3JV43 U4UCM5 A0A1Y1K9C2 A0A1W4WQY7 D6WMM4 A0A023ESP1 N6U883 A0A2J7RJY2 A0A084WJ30 A0A2Z5RE60 Q16P20 U5EUY0 A0A182PV63 Q17GM7 V5G175 A0A1I8JTB6 A0A182UVT4 A0A182KTE7 Q7PV48 A0A182RSS8 A0A336M4V0 A0A182N3L6 A0A182Q2D7 A0A1Y9IVZ9 A0A1Y9J0F6 T1E9V8 A0A2M3Z0V5 T1E2L3 A0A2M4A7U9 A0A2M4A7S6 A0A2M4CUN3 A0A1Q3FUN5 A0A1W5YLL6 A0A182K1V2 A0A182FVT2 A0A182GKM3 A0A182YCD2 A0A1J1IJG4 A0A3Q0IKK7 A0A182M257 A0A1Q3FV06 A0A182JN40 W5J9I7 A0A1W6EVT2 A0A3L8D9H6 A0A1B6LTM4 A0A1B6ERI7 A0A1S3CUV0 A0A1B6FW63 A0A0K8TSU1 A0A1L8E0W5 A0A1L8E196 A0A158NK90 A0A1L8E0T5 A0A1L8EE92 A0A067RFE0 A0A1L8EEN0 T1PDY8 B4N2K9 A0A069DTW1 A0A0P4VSB3 R4G5K7 A0A1B0D970 A0A0V0G5W9 A0A1B6CDS0 A0A224XBZ6 A0A0A1XDV0 A0A0J7NLK9 V5SJ12 A0A195FBS5 D3TQS9 A0A0P4WHU5 Q0QHL3 A0A1I8PUP1 W8CC68 A0A0A9VZC2 A0A023F8N9 R4WJI6 E2BRV0 A0A026X2M5 A0A0P8XS56 A0A151X6U5 A0A1W4VF19 A0A1I9WLM9 E2AVF4 C6TP50 B3NXS1 A0A0M5JAP7 A0A1B6JC96 A0A0R3NYW2 E0VRF1 Q9W401 B4L2G7
PDB
5UQS
E-value=0,
Score=1868
Ontologies
KEGG
PATHWAY
00020
Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01210 2-Oxocarboxylic acid metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
Subcellular location
Mitochondrion matrix
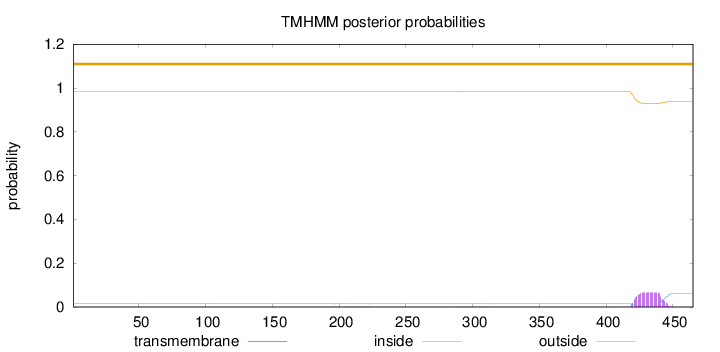
Length:
465
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.46415
Exp number, first 60 AAs:
0.00022
Total prob of N-in:
0.01455
outside
1 - 465
Population Genetic Test Statistics
Pi
19.90474
Theta
16.527872
Tajima's D
0.181385
CLR
0
CSRT
0.421228938553072
Interpretation
Uncertain
