Pre Gene Modal
BGIBMGA000549
Annotation
Putative_serine_protease_K12H4.7_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.079
Sequence
CDS
ATGAAGCTCTACACTATTTTATTTAATTTATATGTAGCGCTAATTAGTGTTGATGGTGTAAAAAAATTTCATTTGGGCCGTAGTAATGGTGGTAATTTAGGAATACCTGGTGGTGATTATCAATCAAATTTGCCACCTCCACAGTGGTTTAAGCAAAAGTTAGACCACTCAAATCCATCCGATCTTCGAACTTGGAAACAGCGCTATTATGTAAATGACAGTTTTTATGATTTCAAAAATCAAGGCCCAGTATTTTTGATGATTGGTGGTGAGGGTCCAGCTGATGCAAGATGGATGGTGACAGGAACTTGGATTAATTATGCAAAGAAGTTCAATGCACTTTGTATAAATTTAGAACATCGTTTTTATGGAGAGAGTCATCCCACTTTGGATCTTAGTATTAAGAACTTACAGTTTCTATCATCATACCAAGCACTTGCTGATCTGGCCAATTTTATCAGTTCAATGAAACAGAAGTTCAGATTAAATGAAAAGGTGAAATGGATTGCCTTTGGAGGATCCTATCCAGGTTCCTTGGCTGCTTGGCTAAGATTAAAATATCCTCATCTTATACATGCATCTATTTCTACAAGTGGACCACTATTGGCTAAAGTTGATTTTAAGGAATATTATCAAGTAGTTGTTGATGCATTAAGAGAAAAGACTGGTGATGATAAATGTGTCAATGAACTTAGGCAAGCGCATAATGAAATAAGTCAATTAATACAACACAGTCCAGAAGTAATTGAAAAGGAATTTAGGGTTTGTAAGCCATTTGGCTTGGCATCCCAAAATGATATGAAAAATTTCTACAATTCAATAGCTGATGATTTTGCTGATTTGGTGCAATACAATGAAGATAACAGGATAAGTGCAGATGTTAATTATAAGAATCTCACAATCAATACAGTTTGCGACATGCTAACAGCAACTGGAGGCTTACCTGCTTACAAGAAACTAGCTGCGTTCAATGATATTGTTCTAGCTAAATCTAACGAAACTTGTATGGATTATAGCTACGATAATATGATCTCAGATCTCAGGAATATAACATGGAGCTCTAATGGAGCTCGGCAATGGATGTATCAAACGTGTACAGAATTCGGATTCTATCAAACGTCATCTGCCGAAATGGATGTCTTTGGAAATAATTTCGATGTCGATTTCTTCATCCAGCAATGTCAAGATGTGTTTGGACAAAAGTACAATCTCAATTTCGTTTCGAACTCCGCTGCATGGACTAACAATTACTATGGTGCTCTAAAAATAGCTGTGGGTCGGATAGTGTTCGTACACGGTTCCATAGATCCGTGGCACGCTCTGGGTATAACCGAGACGAAGGATAACGATTCCCCTGCTATTTTCATACACGGAACCGCACACTGTGCCAATATGTATCCAGCCAGCGATAATGATTTAGCTGAACTGAAACAAGCAAGAATCGAAATTGAAAAATATCTATCCAAATGGCTTGATTGA
Protein
MKLYTILFNLYVALISVDGVKKFHLGRSNGGNLGIPGGDYQSNLPPPQWFKQKLDHSNPSDLRTWKQRYYVNDSFYDFKNQGPVFLMIGGEGPADARWMVTGTWINYAKKFNALCINLEHRFYGESHPTLDLSIKNLQFLSSYQALADLANFISSMKQKFRLNEKVKWIAFGGSYPGSLAAWLRLKYPHLIHASISTSGPLLAKVDFKEYYQVVVDALREKTGDDKCVNELRQAHNEISQLIQHSPEVIEKEFRVCKPFGLASQNDMKNFYNSIADDFADLVQYNEDNRISADVNYKNLTINTVCDMLTATGGLPAYKKLAAFNDIVLAKSNETCMDYSYDNMISDLRNITWSSNGARQWMYQTCTEFGFYQTSSAEMDVFGNNFDVDFFIQQCQDVFGQKYNLNFVSNSAAWTNNYYGALKIAVGRIVFVHGSIDPWHALGITETKDNDSPAIFIHGTAHCANMYPASDNDLAELKQARIEIEKYLSKWLD
Summary
Uniprot
H9ITG9
A0A2H1VNZ5
A0A194PRQ7
A0A194R075
A0A212ETI8
A0A2A4JNU2
+ More
A0A1Y1MRE5 D6WGL2 E9J3M5 A0A151XFV2 A0A158NFD3 A0A195E9L6 A0A151JZ82 F4WUJ0 R4WCS8 A0A195CTE8 A0A087ZYG3 A0A0T6AW67 A0A224XNZ5 A0A2A3E481 A0A1B6JRB9 A0A0C9R157 J9JUF0 T1I703 R4G7X4 A0A2G8JM64 A0A336MJK8 A0A2K8JMA2 A0A3L8D7G5 A0A1I9WL59 A0A2H8TXB1 A0A310SNV3 B4LF33 A0A026WIN4 A0A026VVD7 A0A1B6H0P6 A0A1B6MS95 A0A1V1FNB5 A0A1J1J460 A0A336K475 U5EW69 A0A232EXG9 A0A3L8D5Q3 A0A026VW46 A0A0L7R2V1 A0A2J7QIH2 B4KZU7 A0A067RFD1 A0A1B6CAA8 X1WJF0 B3M9F4 A0A0M3QW94 A0A146KY57 E0VXG1 Q29F35 B4HAY1 A0A0A1WEF6 B0WJZ7 B4MKT7 C3ZCY8 A0A1S3HJ51 A0A1B6I6W3 A0A1S3K8X9 A0A182JP50 U4ULK6 Q9VS02 E2C1R7 A0A0J9RRQ1 B4HV01 B4QJV6 A0A0K8UX52 A0A034WBZ8 B4J384 Q8SZM1 B4PJT8 E9FVW6 A0A2C9K4W9 T1PCZ9 W8BJ07 B3NFK3 A0A1I8PZQ4 A0A182N8C7 A0A182H9D8 A0A182MDR6 A0A023ETD4 A0A1W4V9L3 A0A0L0CRF0 A0A182FD62 A0A0N7ZA73 A0A3B0K479 E2A6B9 A0A182X6X5 A0A084W2L9 W5JI43 A0A1S4FLB2 A0A182WK05 A0A182UJA6 Q7Q6D1 Q16X18 A0A182JL22
A0A1Y1MRE5 D6WGL2 E9J3M5 A0A151XFV2 A0A158NFD3 A0A195E9L6 A0A151JZ82 F4WUJ0 R4WCS8 A0A195CTE8 A0A087ZYG3 A0A0T6AW67 A0A224XNZ5 A0A2A3E481 A0A1B6JRB9 A0A0C9R157 J9JUF0 T1I703 R4G7X4 A0A2G8JM64 A0A336MJK8 A0A2K8JMA2 A0A3L8D7G5 A0A1I9WL59 A0A2H8TXB1 A0A310SNV3 B4LF33 A0A026WIN4 A0A026VVD7 A0A1B6H0P6 A0A1B6MS95 A0A1V1FNB5 A0A1J1J460 A0A336K475 U5EW69 A0A232EXG9 A0A3L8D5Q3 A0A026VW46 A0A0L7R2V1 A0A2J7QIH2 B4KZU7 A0A067RFD1 A0A1B6CAA8 X1WJF0 B3M9F4 A0A0M3QW94 A0A146KY57 E0VXG1 Q29F35 B4HAY1 A0A0A1WEF6 B0WJZ7 B4MKT7 C3ZCY8 A0A1S3HJ51 A0A1B6I6W3 A0A1S3K8X9 A0A182JP50 U4ULK6 Q9VS02 E2C1R7 A0A0J9RRQ1 B4HV01 B4QJV6 A0A0K8UX52 A0A034WBZ8 B4J384 Q8SZM1 B4PJT8 E9FVW6 A0A2C9K4W9 T1PCZ9 W8BJ07 B3NFK3 A0A1I8PZQ4 A0A182N8C7 A0A182H9D8 A0A182MDR6 A0A023ETD4 A0A1W4V9L3 A0A0L0CRF0 A0A182FD62 A0A0N7ZA73 A0A3B0K479 E2A6B9 A0A182X6X5 A0A084W2L9 W5JI43 A0A1S4FLB2 A0A182WK05 A0A182UJA6 Q7Q6D1 Q16X18 A0A182JL22
Pubmed
19121390
26354079
22118469
28004739
18362917
19820115
+ More
21282665 21347285 21719571 23691247 29023486 30249741 27538518 17994087 24508170 28410430 28648823 24845553 26823975 20566863 15632085 25830018 18563158 26383154 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 22936249 25348373 17550304 21292972 15562597 25315136 24495485 26483478 24945155 26108605 24438588 20920257 23761445 17510324 12364791 14747013 17210077
21282665 21347285 21719571 23691247 29023486 30249741 27538518 17994087 24508170 28410430 28648823 24845553 26823975 20566863 15632085 25830018 18563158 26383154 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20798317 22936249 25348373 17550304 21292972 15562597 25315136 24495485 26483478 24945155 26108605 24438588 20920257 23761445 17510324 12364791 14747013 17210077
EMBL
BABH01006977
BABH01006978
ODYU01003579
SOQ42517.1
KQ459594
KPI96131.1
+ More
KQ461108 KPJ09251.1 AGBW02012593 OWR44761.1 NWSH01000884 PCG73717.1 GEZM01025029 JAV87638.1 KQ971321 EFA00580.2 GL768066 EFZ12594.1 KQ982182 KYQ59185.1 ADTU01014274 KQ979440 KYN21502.1 KQ981410 KYN41918.1 GL888365 EGI62155.1 AK417138 BAN20353.1 KQ977279 KYN03978.1 LJIG01022644 KRT79472.1 GFTR01006675 JAW09751.1 KZ288400 PBC26304.1 GECU01005922 JAT01785.1 GBYB01000596 JAG70363.1 ABLF02028806 ABLF02028809 ACPB03000772 GAHY01001967 JAA75543.1 MRZV01001613 PIK36813.1 UFQT01001440 SSX30592.1 KY031188 ATU82939.1 QOIP01000012 RLU16239.1 KU932243 APA33879.1 GFXV01006003 MBW17808.1 KQ762408 OAD55889.1 CH940647 EDW69202.1 KK107199 EZA55516.1 KK108267 EZA46799.1 GECZ01001513 JAS68256.1 GEBQ01001166 JAT38811.1 FX985297 BAX07310.1 CVRI01000067 CRL06754.1 UFQS01000109 UFQT01000109 SSW99774.1 SSX20154.1 GANO01003087 JAB56784.1 NNAY01001721 OXU23126.1 QOIP01000013 RLU15564.1 KK107796 EZA47726.1 KQ414665 KOC65159.1 NEVH01013592 PNF28388.1 CH933809 EDW17959.1 KK852542 KDR21723.1 GEDC01026876 JAS10422.1 ABLF02021908 ABLF02021912 CH902618 EDV41167.1 CP012525 ALC43759.1 GDHC01017256 JAQ01373.1 DS235832 EEB18067.1 CH379068 EAL31237.1 CH479246 EDW37775.1 GBXI01016903 JAC97388.1 DS231966 EDS29541.1 CH963847 EDW72793.1 GG666610 EEN49641.1 GECU01025044 JAS82662.1 KB632273 ERL91071.1 AE014296 AAF50628.1 GL451975 EFN78126.1 CM002912 KMY97979.1 CH480817 EDW50772.1 CM000363 EDX09497.1 GDHF01021092 GDHF01011848 JAI31222.1 JAI40466.1 GAKP01007297 GAKP01007296 JAC51655.1 CH916366 EDV96155.1 AY070659 AAL48130.1 CM000159 EDW93687.1 GL732525 EFX88622.1 KA646544 AFP61173.1 GAMC01009592 JAB96963.1 CH954178 EDV50545.1 JXUM01120459 KQ566394 KXJ70163.1 AXCM01000891 GAPW01001040 JAC12558.1 JRES01000122 KNC34009.1 GDRN01103633 JAI58048.1 OUUW01000012 SPP87512.1 GL437123 EFN71004.1 ATLV01019633 KE525275 KFB44463.1 ADMH02001182 ETN63801.1 AAAB01008960 EAA11647.3 CH477548 EAT39136.1
KQ461108 KPJ09251.1 AGBW02012593 OWR44761.1 NWSH01000884 PCG73717.1 GEZM01025029 JAV87638.1 KQ971321 EFA00580.2 GL768066 EFZ12594.1 KQ982182 KYQ59185.1 ADTU01014274 KQ979440 KYN21502.1 KQ981410 KYN41918.1 GL888365 EGI62155.1 AK417138 BAN20353.1 KQ977279 KYN03978.1 LJIG01022644 KRT79472.1 GFTR01006675 JAW09751.1 KZ288400 PBC26304.1 GECU01005922 JAT01785.1 GBYB01000596 JAG70363.1 ABLF02028806 ABLF02028809 ACPB03000772 GAHY01001967 JAA75543.1 MRZV01001613 PIK36813.1 UFQT01001440 SSX30592.1 KY031188 ATU82939.1 QOIP01000012 RLU16239.1 KU932243 APA33879.1 GFXV01006003 MBW17808.1 KQ762408 OAD55889.1 CH940647 EDW69202.1 KK107199 EZA55516.1 KK108267 EZA46799.1 GECZ01001513 JAS68256.1 GEBQ01001166 JAT38811.1 FX985297 BAX07310.1 CVRI01000067 CRL06754.1 UFQS01000109 UFQT01000109 SSW99774.1 SSX20154.1 GANO01003087 JAB56784.1 NNAY01001721 OXU23126.1 QOIP01000013 RLU15564.1 KK107796 EZA47726.1 KQ414665 KOC65159.1 NEVH01013592 PNF28388.1 CH933809 EDW17959.1 KK852542 KDR21723.1 GEDC01026876 JAS10422.1 ABLF02021908 ABLF02021912 CH902618 EDV41167.1 CP012525 ALC43759.1 GDHC01017256 JAQ01373.1 DS235832 EEB18067.1 CH379068 EAL31237.1 CH479246 EDW37775.1 GBXI01016903 JAC97388.1 DS231966 EDS29541.1 CH963847 EDW72793.1 GG666610 EEN49641.1 GECU01025044 JAS82662.1 KB632273 ERL91071.1 AE014296 AAF50628.1 GL451975 EFN78126.1 CM002912 KMY97979.1 CH480817 EDW50772.1 CM000363 EDX09497.1 GDHF01021092 GDHF01011848 JAI31222.1 JAI40466.1 GAKP01007297 GAKP01007296 JAC51655.1 CH916366 EDV96155.1 AY070659 AAL48130.1 CM000159 EDW93687.1 GL732525 EFX88622.1 KA646544 AFP61173.1 GAMC01009592 JAB96963.1 CH954178 EDV50545.1 JXUM01120459 KQ566394 KXJ70163.1 AXCM01000891 GAPW01001040 JAC12558.1 JRES01000122 KNC34009.1 GDRN01103633 JAI58048.1 OUUW01000012 SPP87512.1 GL437123 EFN71004.1 ATLV01019633 KE525275 KFB44463.1 ADMH02001182 ETN63801.1 AAAB01008960 EAA11647.3 CH477548 EAT39136.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000218220
UP000007266
+ More
UP000075809 UP000005205 UP000078492 UP000078541 UP000007755 UP000078542 UP000005203 UP000242457 UP000007819 UP000015103 UP000230750 UP000279307 UP000008792 UP000053097 UP000183832 UP000215335 UP000053825 UP000235965 UP000009192 UP000027135 UP000007801 UP000092553 UP000009046 UP000001819 UP000008744 UP000002320 UP000007798 UP000001554 UP000085678 UP000075881 UP000030742 UP000000803 UP000008237 UP000001292 UP000000304 UP000001070 UP000002282 UP000000305 UP000076420 UP000095301 UP000008711 UP000095300 UP000075884 UP000069940 UP000249989 UP000075883 UP000192221 UP000037069 UP000069272 UP000268350 UP000000311 UP000076407 UP000030765 UP000000673 UP000075920 UP000075902 UP000007062 UP000008820 UP000075880
UP000075809 UP000005205 UP000078492 UP000078541 UP000007755 UP000078542 UP000005203 UP000242457 UP000007819 UP000015103 UP000230750 UP000279307 UP000008792 UP000053097 UP000183832 UP000215335 UP000053825 UP000235965 UP000009192 UP000027135 UP000007801 UP000092553 UP000009046 UP000001819 UP000008744 UP000002320 UP000007798 UP000001554 UP000085678 UP000075881 UP000030742 UP000000803 UP000008237 UP000001292 UP000000304 UP000001070 UP000002282 UP000000305 UP000076420 UP000095301 UP000008711 UP000095300 UP000075884 UP000069940 UP000249989 UP000075883 UP000192221 UP000037069 UP000069272 UP000268350 UP000000311 UP000076407 UP000030765 UP000000673 UP000075920 UP000075902 UP000007062 UP000008820 UP000075880
PRIDE
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9ITG9
A0A2H1VNZ5
A0A194PRQ7
A0A194R075
A0A212ETI8
A0A2A4JNU2
+ More
A0A1Y1MRE5 D6WGL2 E9J3M5 A0A151XFV2 A0A158NFD3 A0A195E9L6 A0A151JZ82 F4WUJ0 R4WCS8 A0A195CTE8 A0A087ZYG3 A0A0T6AW67 A0A224XNZ5 A0A2A3E481 A0A1B6JRB9 A0A0C9R157 J9JUF0 T1I703 R4G7X4 A0A2G8JM64 A0A336MJK8 A0A2K8JMA2 A0A3L8D7G5 A0A1I9WL59 A0A2H8TXB1 A0A310SNV3 B4LF33 A0A026WIN4 A0A026VVD7 A0A1B6H0P6 A0A1B6MS95 A0A1V1FNB5 A0A1J1J460 A0A336K475 U5EW69 A0A232EXG9 A0A3L8D5Q3 A0A026VW46 A0A0L7R2V1 A0A2J7QIH2 B4KZU7 A0A067RFD1 A0A1B6CAA8 X1WJF0 B3M9F4 A0A0M3QW94 A0A146KY57 E0VXG1 Q29F35 B4HAY1 A0A0A1WEF6 B0WJZ7 B4MKT7 C3ZCY8 A0A1S3HJ51 A0A1B6I6W3 A0A1S3K8X9 A0A182JP50 U4ULK6 Q9VS02 E2C1R7 A0A0J9RRQ1 B4HV01 B4QJV6 A0A0K8UX52 A0A034WBZ8 B4J384 Q8SZM1 B4PJT8 E9FVW6 A0A2C9K4W9 T1PCZ9 W8BJ07 B3NFK3 A0A1I8PZQ4 A0A182N8C7 A0A182H9D8 A0A182MDR6 A0A023ETD4 A0A1W4V9L3 A0A0L0CRF0 A0A182FD62 A0A0N7ZA73 A0A3B0K479 E2A6B9 A0A182X6X5 A0A084W2L9 W5JI43 A0A1S4FLB2 A0A182WK05 A0A182UJA6 Q7Q6D1 Q16X18 A0A182JL22
A0A1Y1MRE5 D6WGL2 E9J3M5 A0A151XFV2 A0A158NFD3 A0A195E9L6 A0A151JZ82 F4WUJ0 R4WCS8 A0A195CTE8 A0A087ZYG3 A0A0T6AW67 A0A224XNZ5 A0A2A3E481 A0A1B6JRB9 A0A0C9R157 J9JUF0 T1I703 R4G7X4 A0A2G8JM64 A0A336MJK8 A0A2K8JMA2 A0A3L8D7G5 A0A1I9WL59 A0A2H8TXB1 A0A310SNV3 B4LF33 A0A026WIN4 A0A026VVD7 A0A1B6H0P6 A0A1B6MS95 A0A1V1FNB5 A0A1J1J460 A0A336K475 U5EW69 A0A232EXG9 A0A3L8D5Q3 A0A026VW46 A0A0L7R2V1 A0A2J7QIH2 B4KZU7 A0A067RFD1 A0A1B6CAA8 X1WJF0 B3M9F4 A0A0M3QW94 A0A146KY57 E0VXG1 Q29F35 B4HAY1 A0A0A1WEF6 B0WJZ7 B4MKT7 C3ZCY8 A0A1S3HJ51 A0A1B6I6W3 A0A1S3K8X9 A0A182JP50 U4ULK6 Q9VS02 E2C1R7 A0A0J9RRQ1 B4HV01 B4QJV6 A0A0K8UX52 A0A034WBZ8 B4J384 Q8SZM1 B4PJT8 E9FVW6 A0A2C9K4W9 T1PCZ9 W8BJ07 B3NFK3 A0A1I8PZQ4 A0A182N8C7 A0A182H9D8 A0A182MDR6 A0A023ETD4 A0A1W4V9L3 A0A0L0CRF0 A0A182FD62 A0A0N7ZA73 A0A3B0K479 E2A6B9 A0A182X6X5 A0A084W2L9 W5JI43 A0A1S4FLB2 A0A182WK05 A0A182UJA6 Q7Q6D1 Q16X18 A0A182JL22
PDB
3N0T
E-value=8.45149e-34,
Score=360
Ontologies
GO
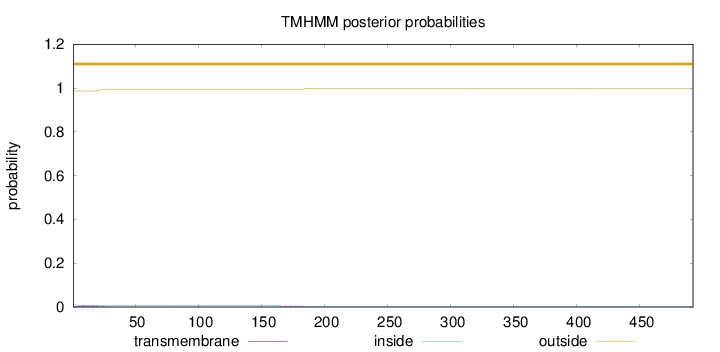
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.952699
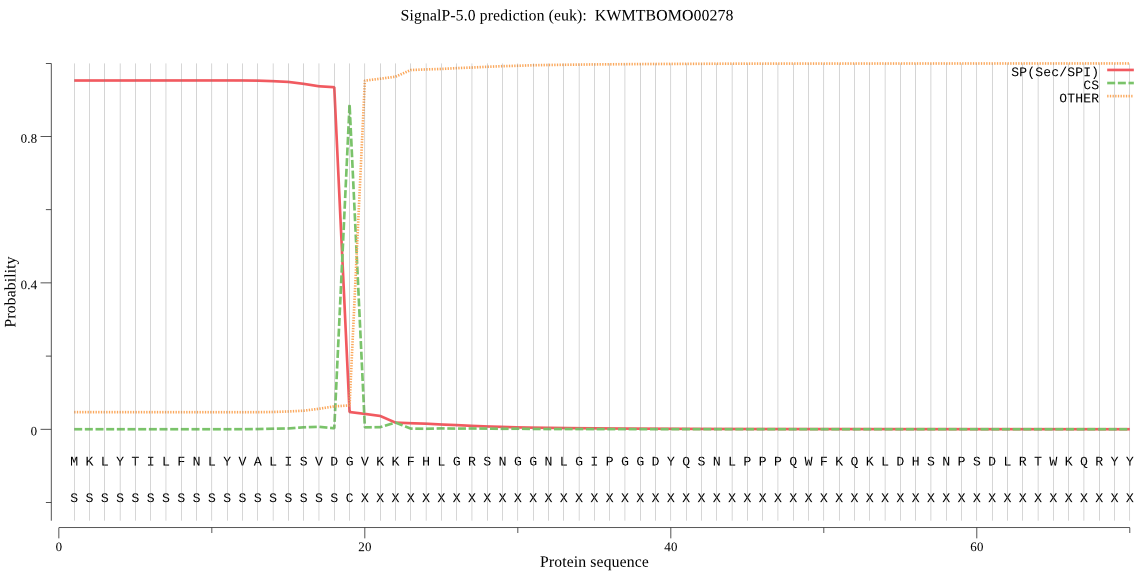
Length:
492
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24704
Exp number, first 60 AAs:
0.12973
Total prob of N-in:
0.01323
outside
1 - 492
Population Genetic Test Statistics
Pi
21.712913
Theta
20.536693
Tajima's D
-1.200649
CLR
0.126742
CSRT
0.101694915254237
Interpretation
Uncertain
