Gene
KWMTBOMO00277
Pre Gene Modal
BGIBMGA000671
Annotation
PREDICTED:_proton-coupled_folate_transporter_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.884
Sequence
CDS
ATGTCAACTACAAGCAAAGAAAATATAGGCATGATTCAACCAAGGATGCCATGGTACAAATATATAACGGTGGAACCAACAATGTTCTTCTACATGATGGCATATATGATCACAAATGTAATAGAGCAAACATTTTATGTCTTTCAAACGTGTCATATTAATCATGGCTATAGTACAGAAATATGTTACAACATTAGCAAACATGCAGATATCAATAAAGAAGTACAGGTGACAGTATCAACATTCCATCAATGGAACGGTATAGCAAGTCATGTTGTTCCTCTATTTCTAGCCTTTTTCTTGGGCTCATACAGTGATAAAAGAGGACGAAAAATTGTATTATTGGCTGGGCTACTGGGAAAATTATATTTTTCCATCATGATTACTGTGAATACAATGAATGATTGGCCTGTAGAATATGTGATATACACGGCTGCTCTACCAAGTGCACTGACCGGTGCTGACTTGGCTATATTTGCCGGATGTTTTGCTTACATAGCAGATGTATCTTCAGTAAAAAATAGAACATTGCGTGTTGGAATATTAGACGTAACTTATTTAAGCACATTGCCCATGGGTATAGCTATAGGAAATTTATTATACAATAAGGTAGTAAATAAGTCGTTCACGTCAATGTTTGCAATCAATACATTTCTAATGGTTCTGGCGACGTTACATTGTTTAATATTTCTCGATTGGCAAACACGTCAAGAACAGAAGTCTCTAAAAGAAGCCGGCGTAAAGAACCCGTTTACTGATTTCTTCGACACAAATAATATCGTTCAGACTGTTGTTACGTTAACAAGAAGACGATCGAACAATAAAAGACTATTTTTGTGGCTTTTGCTTACATCGATGGCATTCTATACATTTCAAAGAGATGAAAAACAAGTGATGTACTTATATGCACAGACCGTCTTCAAATGGAACGAAACCACATATAGCAACTTTAAAACGTACTTGAGCACATTGTATGTGATTGCTATGTTGTTTGGTATACCTTTAATGACTAAACTACTGAAGTGGAGGGACACGTTAATAGTGATGCTTGGTGCCGCAGCCCACATAACCGGGCATCTAGTGTATGCTCACACCACAGTTGGAAAGATGATGTACGTGGGTGCTACGGCGGCGGCATTAGGACCCATTGTGGCACCACTCATACGTTCCATTACTTCAAAACTGTTACCGCCTGATGAAAGAGGTGTAGCATACGCTTTCTTATCTGTCATGGAGAACGCAGTAGCTATATTCGCGTCGATCGTGTATACGCAGATTTATAATGCTACCATCGGCACGGAATACATAAATTCAATATTTTATTTTACTATATCGACGCAAGTTATAGTTTTTCTAAGTGCTTTCACAATGGATATATTGTTAAAAGGCAAAAAATTTGATGATATTAGTAACGAAGAAGAGAAAAGTATTGCCATGAGTTAA
Protein
MSTTSKENIGMIQPRMPWYKYITVEPTMFFYMMAYMITNVIEQTFYVFQTCHINHGYSTEICYNISKHADINKEVQVTVSTFHQWNGIASHVVPLFLAFFLGSYSDKRGRKIVLLAGLLGKLYFSIMITVNTMNDWPVEYVIYTAALPSALTGADLAIFAGCFAYIADVSSVKNRTLRVGILDVTYLSTLPMGIAIGNLLYNKVVNKSFTSMFAINTFLMVLATLHCLIFLDWQTRQEQKSLKEAGVKNPFTDFFDTNNIVQTVVTLTRRRSNNKRLFLWLLLTSMAFYTFQRDEKQVMYLYAQTVFKWNETTYSNFKTYLSTLYVIAMLFGIPLMTKLLKWRDTLIVMLGAAAHITGHLVYAHTTVGKMMYVGATAAALGPIVAPLIRSITSKLLPPDERGVAYAFLSVMENAVAIFASIVYTQIYNATIGTEYINSIFYFTISTQVIVFLSAFTMDILLKGKKFDDISNEEEKSIAMS
Summary
Uniprot
A0A212ETE0
A0A2A4JPT6
A0A194PYA4
A0A194QUV7
Q8JKI7
G9I049
+ More
A0A0L7LKJ6 A0A1L8DED7 W5J2J2 T1DU24 A0A182JG93 A0A2M4BK94 A0A2M4BKB4 A0A182FRD2 A0A182LFV8 A0A182WRL5 A0A182TXI3 A0A182V760 A0A182HU63 A0A084VNP6 A0A182RNT0 A0A182N655 Q7PGR0 A0A182LXU4 A0A182Y492 A0A182QDM3 A0A182W047 A0A182SHH0 Q17ME2 A0A2M4BM70 A0A182PKM3 A0A1L8DEA3 U5ETF5 A0A182JXV0 A0A336MAY0 A0A336KVN1 A0A1J1IHV4 B0WJS7 D6WVQ3 A0A1Q3FI83 A0A182HDQ2 A0A139WDN3 T1PIP6 A0A1I8MCY8 A0A0K8V6M0 W8BGL2 A0A1I8PBN0 A0A2A3ED49 V9IBM7 A0A0K8UZJ5 A0A0K8UFG8 A0A0A1WCU8 A0A0A1XA55 A0A0C9QHP5 B4MMH5 A0A0Q9WPF2 A0A1W4WV72 A0A0L0C1X3 A0A0Q9XS21 B4L9J2 A0A0Q9XHH0 A0A1A9YSS3 A0A1B0A6Z5 A0A1B0FJU6 A0A1B0B6N5 B4PKE1 A0A1A9W3A1 A0A2J7RA40 A0A151X6Z3 A0A1A9UIF7 Q0E8E1 Q9I7R2 Q960A8 F4W8H4 A0A0M4EZ89 B4J0W1 E1ZZU3 A0A0L7R0X9 B4LI51 A0A0J9RWU5 A0A0Q9WPH0 A0A0Q9WH72 A0A195FAJ5 A0A0J9RWL4 B4HJN8 B3NHZ4 A0A195BAW8 A0A1W4UGF7 A0A0K8W0D7 A0A026WU70 A0A195CYM9 A0A195EHS8 A0A158NK79 B3M4H9 A0A0R1DYU9 B4H219 A0A0R3P8G5 A0A0R3P6L1 B5DRB4
A0A0L7LKJ6 A0A1L8DED7 W5J2J2 T1DU24 A0A182JG93 A0A2M4BK94 A0A2M4BKB4 A0A182FRD2 A0A182LFV8 A0A182WRL5 A0A182TXI3 A0A182V760 A0A182HU63 A0A084VNP6 A0A182RNT0 A0A182N655 Q7PGR0 A0A182LXU4 A0A182Y492 A0A182QDM3 A0A182W047 A0A182SHH0 Q17ME2 A0A2M4BM70 A0A182PKM3 A0A1L8DEA3 U5ETF5 A0A182JXV0 A0A336MAY0 A0A336KVN1 A0A1J1IHV4 B0WJS7 D6WVQ3 A0A1Q3FI83 A0A182HDQ2 A0A139WDN3 T1PIP6 A0A1I8MCY8 A0A0K8V6M0 W8BGL2 A0A1I8PBN0 A0A2A3ED49 V9IBM7 A0A0K8UZJ5 A0A0K8UFG8 A0A0A1WCU8 A0A0A1XA55 A0A0C9QHP5 B4MMH5 A0A0Q9WPF2 A0A1W4WV72 A0A0L0C1X3 A0A0Q9XS21 B4L9J2 A0A0Q9XHH0 A0A1A9YSS3 A0A1B0A6Z5 A0A1B0FJU6 A0A1B0B6N5 B4PKE1 A0A1A9W3A1 A0A2J7RA40 A0A151X6Z3 A0A1A9UIF7 Q0E8E1 Q9I7R2 Q960A8 F4W8H4 A0A0M4EZ89 B4J0W1 E1ZZU3 A0A0L7R0X9 B4LI51 A0A0J9RWU5 A0A0Q9WPH0 A0A0Q9WH72 A0A195FAJ5 A0A0J9RWL4 B4HJN8 B3NHZ4 A0A195BAW8 A0A1W4UGF7 A0A0K8W0D7 A0A026WU70 A0A195CYM9 A0A195EHS8 A0A158NK79 B3M4H9 A0A0R1DYU9 B4H219 A0A0R3P8G5 A0A0R3P6L1 B5DRB4
Pubmed
22118469
26354079
12186886
22355451
26227816
20920257
+ More
23761445 20966253 24438588 12364791 25244985 17510324 18362917 19820115 26483478 25315136 24495485 25830018 17994087 26108605 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20798317 22936249 24508170 30249741 21347285 15632085
23761445 20966253 24438588 12364791 25244985 17510324 18362917 19820115 26483478 25315136 24495485 25830018 17994087 26108605 18057021 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 20798317 22936249 24508170 30249741 21347285 15632085
EMBL
AGBW02012593
OWR44758.1
NWSH01000884
PCG73718.1
KQ459594
KPI96130.1
+ More
KQ461108 KPJ09252.1 AF451898 AAN04419.1 JN418988 AEW69572.1 JTDY01000766 KOB75955.1 GFDF01009364 JAV04720.1 ADMH02002131 ETN58482.1 GAMD01000539 JAB01052.1 GGFJ01004358 MBW53499.1 GGFJ01004359 MBW53500.1 APCN01000608 ATLV01014811 KE524989 KFB39590.1 AAAB01008859 EAA44839.5 AXCM01000791 AXCN02000196 CH477206 EAT47896.1 GGFJ01004900 MBW54041.1 GFDF01009404 JAV04680.1 GANO01001943 JAB57928.1 UFQT01000586 SSX25517.1 UFQS01000990 UFQT01000990 SSX08295.1 SSX28318.1 CVRI01000053 CRK99795.1 DS231963 EDS29420.1 KQ971358 EFA08602.2 GFDL01007754 JAV27291.1 JXUM01130112 JXUM01130113 KQ567538 KXJ69387.1 KYB25977.1 KA647808 AFP62437.1 GDHF01029661 GDHF01017730 GDHF01016157 GDHF01003664 JAI22653.1 JAI34584.1 JAI36157.1 JAI48650.1 GAMC01017759 GAMC01017757 GAMC01017756 JAB88799.1 KZ288280 PBC29657.1 JR037318 AEY57709.1 GDHF01026685 GDHF01020283 JAI25629.1 JAI32031.1 GDHF01026925 JAI25389.1 GBXI01017580 JAC96711.1 GBXI01006739 JAD07553.1 GBYB01000092 GBYB01006701 JAG69859.1 JAG76468.1 CH963847 EDW73320.2 KRF97744.1 JRES01000997 KNC26257.1 CH933816 KRG07758.1 EDW17367.2 KRG07757.1 KRG07759.1 KRG07760.1 CCAG010018840 JXJN01009178 CM000159 EDW94839.1 KRK02080.1 KRK02081.1 NEVH01006578 PNF37708.1 KQ982472 KYQ56099.1 AE014296 BT100042 AAF49456.1 AAG22317.1 ACX61583.1 AAG22318.1 AY052147 AAK93571.1 GL887908 EGI69465.1 CP012525 ALC43573.1 ALC43987.1 ALC44845.1 ALC44851.1 CH916366 EDV95782.1 GL435522 EFN73316.1 KQ414670 KOC64451.1 CH940647 EDW68595.1 KRF83910.1 KRF83912.1 CM002912 KMZ00058.1 KMZ00059.1 KRF83911.1 KRF83913.1 KQ981693 KYN37645.1 KMZ00057.1 CH480815 EDW41765.1 CH954178 EDV52080.2 KQ976537 KYM81365.1 GDHF01007712 JAI44602.1 KK107105 QOIP01000011 EZA59582.1 RLU17053.1 KQ977110 KYN05682.1 KQ978881 KYN27808.1 ADTU01018731 CH902618 EDV40473.1 KPU78581.1 KRK02079.1 CH479203 EDW30371.1 CH379070 KRT08639.1 KRT08638.1 EDY74019.1
KQ461108 KPJ09252.1 AF451898 AAN04419.1 JN418988 AEW69572.1 JTDY01000766 KOB75955.1 GFDF01009364 JAV04720.1 ADMH02002131 ETN58482.1 GAMD01000539 JAB01052.1 GGFJ01004358 MBW53499.1 GGFJ01004359 MBW53500.1 APCN01000608 ATLV01014811 KE524989 KFB39590.1 AAAB01008859 EAA44839.5 AXCM01000791 AXCN02000196 CH477206 EAT47896.1 GGFJ01004900 MBW54041.1 GFDF01009404 JAV04680.1 GANO01001943 JAB57928.1 UFQT01000586 SSX25517.1 UFQS01000990 UFQT01000990 SSX08295.1 SSX28318.1 CVRI01000053 CRK99795.1 DS231963 EDS29420.1 KQ971358 EFA08602.2 GFDL01007754 JAV27291.1 JXUM01130112 JXUM01130113 KQ567538 KXJ69387.1 KYB25977.1 KA647808 AFP62437.1 GDHF01029661 GDHF01017730 GDHF01016157 GDHF01003664 JAI22653.1 JAI34584.1 JAI36157.1 JAI48650.1 GAMC01017759 GAMC01017757 GAMC01017756 JAB88799.1 KZ288280 PBC29657.1 JR037318 AEY57709.1 GDHF01026685 GDHF01020283 JAI25629.1 JAI32031.1 GDHF01026925 JAI25389.1 GBXI01017580 JAC96711.1 GBXI01006739 JAD07553.1 GBYB01000092 GBYB01006701 JAG69859.1 JAG76468.1 CH963847 EDW73320.2 KRF97744.1 JRES01000997 KNC26257.1 CH933816 KRG07758.1 EDW17367.2 KRG07757.1 KRG07759.1 KRG07760.1 CCAG010018840 JXJN01009178 CM000159 EDW94839.1 KRK02080.1 KRK02081.1 NEVH01006578 PNF37708.1 KQ982472 KYQ56099.1 AE014296 BT100042 AAF49456.1 AAG22317.1 ACX61583.1 AAG22318.1 AY052147 AAK93571.1 GL887908 EGI69465.1 CP012525 ALC43573.1 ALC43987.1 ALC44845.1 ALC44851.1 CH916366 EDV95782.1 GL435522 EFN73316.1 KQ414670 KOC64451.1 CH940647 EDW68595.1 KRF83910.1 KRF83912.1 CM002912 KMZ00058.1 KMZ00059.1 KRF83911.1 KRF83913.1 KQ981693 KYN37645.1 KMZ00057.1 CH480815 EDW41765.1 CH954178 EDV52080.2 KQ976537 KYM81365.1 GDHF01007712 JAI44602.1 KK107105 QOIP01000011 EZA59582.1 RLU17053.1 KQ977110 KYN05682.1 KQ978881 KYN27808.1 ADTU01018731 CH902618 EDV40473.1 KPU78581.1 KRK02079.1 CH479203 EDW30371.1 CH379070 KRT08639.1 KRT08638.1 EDY74019.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000232784
UP000029779
+ More
UP000037510 UP000000673 UP000075880 UP000069272 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000030765 UP000075900 UP000075884 UP000007062 UP000075883 UP000076408 UP000075886 UP000075920 UP000075901 UP000008820 UP000075885 UP000075881 UP000183832 UP000002320 UP000007266 UP000069940 UP000249989 UP000095301 UP000095300 UP000242457 UP000007798 UP000192223 UP000037069 UP000009192 UP000092443 UP000092445 UP000092444 UP000092460 UP000002282 UP000091820 UP000235965 UP000075809 UP000078200 UP000000803 UP000007755 UP000092553 UP000001070 UP000000311 UP000053825 UP000008792 UP000078541 UP000001292 UP000008711 UP000078540 UP000192221 UP000053097 UP000279307 UP000078542 UP000078492 UP000005205 UP000007801 UP000008744 UP000001819
UP000037510 UP000000673 UP000075880 UP000069272 UP000075882 UP000076407 UP000075902 UP000075903 UP000075840 UP000030765 UP000075900 UP000075884 UP000007062 UP000075883 UP000076408 UP000075886 UP000075920 UP000075901 UP000008820 UP000075885 UP000075881 UP000183832 UP000002320 UP000007266 UP000069940 UP000249989 UP000095301 UP000095300 UP000242457 UP000007798 UP000192223 UP000037069 UP000009192 UP000092443 UP000092445 UP000092444 UP000092460 UP000002282 UP000091820 UP000235965 UP000075809 UP000078200 UP000000803 UP000007755 UP000092553 UP000001070 UP000000311 UP000053825 UP000008792 UP000078541 UP000001292 UP000008711 UP000078540 UP000192221 UP000053097 UP000279307 UP000078542 UP000078492 UP000005205 UP000007801 UP000008744 UP000001819
Pfam
PF07690 MFS_1
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A212ETE0
A0A2A4JPT6
A0A194PYA4
A0A194QUV7
Q8JKI7
G9I049
+ More
A0A0L7LKJ6 A0A1L8DED7 W5J2J2 T1DU24 A0A182JG93 A0A2M4BK94 A0A2M4BKB4 A0A182FRD2 A0A182LFV8 A0A182WRL5 A0A182TXI3 A0A182V760 A0A182HU63 A0A084VNP6 A0A182RNT0 A0A182N655 Q7PGR0 A0A182LXU4 A0A182Y492 A0A182QDM3 A0A182W047 A0A182SHH0 Q17ME2 A0A2M4BM70 A0A182PKM3 A0A1L8DEA3 U5ETF5 A0A182JXV0 A0A336MAY0 A0A336KVN1 A0A1J1IHV4 B0WJS7 D6WVQ3 A0A1Q3FI83 A0A182HDQ2 A0A139WDN3 T1PIP6 A0A1I8MCY8 A0A0K8V6M0 W8BGL2 A0A1I8PBN0 A0A2A3ED49 V9IBM7 A0A0K8UZJ5 A0A0K8UFG8 A0A0A1WCU8 A0A0A1XA55 A0A0C9QHP5 B4MMH5 A0A0Q9WPF2 A0A1W4WV72 A0A0L0C1X3 A0A0Q9XS21 B4L9J2 A0A0Q9XHH0 A0A1A9YSS3 A0A1B0A6Z5 A0A1B0FJU6 A0A1B0B6N5 B4PKE1 A0A1A9W3A1 A0A2J7RA40 A0A151X6Z3 A0A1A9UIF7 Q0E8E1 Q9I7R2 Q960A8 F4W8H4 A0A0M4EZ89 B4J0W1 E1ZZU3 A0A0L7R0X9 B4LI51 A0A0J9RWU5 A0A0Q9WPH0 A0A0Q9WH72 A0A195FAJ5 A0A0J9RWL4 B4HJN8 B3NHZ4 A0A195BAW8 A0A1W4UGF7 A0A0K8W0D7 A0A026WU70 A0A195CYM9 A0A195EHS8 A0A158NK79 B3M4H9 A0A0R1DYU9 B4H219 A0A0R3P8G5 A0A0R3P6L1 B5DRB4
A0A0L7LKJ6 A0A1L8DED7 W5J2J2 T1DU24 A0A182JG93 A0A2M4BK94 A0A2M4BKB4 A0A182FRD2 A0A182LFV8 A0A182WRL5 A0A182TXI3 A0A182V760 A0A182HU63 A0A084VNP6 A0A182RNT0 A0A182N655 Q7PGR0 A0A182LXU4 A0A182Y492 A0A182QDM3 A0A182W047 A0A182SHH0 Q17ME2 A0A2M4BM70 A0A182PKM3 A0A1L8DEA3 U5ETF5 A0A182JXV0 A0A336MAY0 A0A336KVN1 A0A1J1IHV4 B0WJS7 D6WVQ3 A0A1Q3FI83 A0A182HDQ2 A0A139WDN3 T1PIP6 A0A1I8MCY8 A0A0K8V6M0 W8BGL2 A0A1I8PBN0 A0A2A3ED49 V9IBM7 A0A0K8UZJ5 A0A0K8UFG8 A0A0A1WCU8 A0A0A1XA55 A0A0C9QHP5 B4MMH5 A0A0Q9WPF2 A0A1W4WV72 A0A0L0C1X3 A0A0Q9XS21 B4L9J2 A0A0Q9XHH0 A0A1A9YSS3 A0A1B0A6Z5 A0A1B0FJU6 A0A1B0B6N5 B4PKE1 A0A1A9W3A1 A0A2J7RA40 A0A151X6Z3 A0A1A9UIF7 Q0E8E1 Q9I7R2 Q960A8 F4W8H4 A0A0M4EZ89 B4J0W1 E1ZZU3 A0A0L7R0X9 B4LI51 A0A0J9RWU5 A0A0Q9WPH0 A0A0Q9WH72 A0A195FAJ5 A0A0J9RWL4 B4HJN8 B3NHZ4 A0A195BAW8 A0A1W4UGF7 A0A0K8W0D7 A0A026WU70 A0A195CYM9 A0A195EHS8 A0A158NK79 B3M4H9 A0A0R1DYU9 B4H219 A0A0R3P8G5 A0A0R3P6L1 B5DRB4
Ontologies
GO
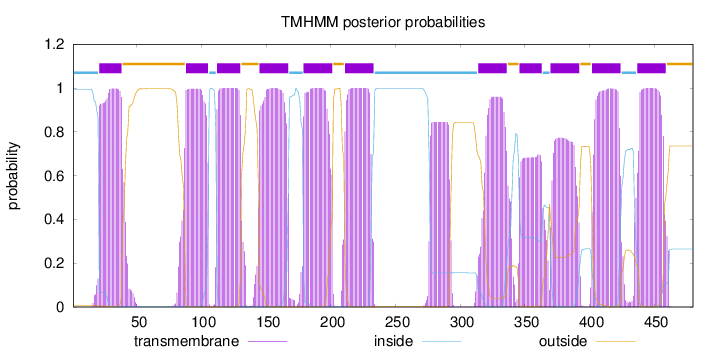
Topology
Length:
480
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
235.27882
Exp number, first 60 AAs:
20.18602
Total prob of N-in:
0.99459
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 38
outside
39 - 87
TMhelix
88 - 105
inside
106 - 111
TMhelix
112 - 130
outside
131 - 144
TMhelix
145 - 167
inside
168 - 178
TMhelix
179 - 201
outside
202 - 210
TMhelix
211 - 233
inside
234 - 313
TMhelix
314 - 336
outside
337 - 345
TMhelix
346 - 363
inside
364 - 369
TMhelix
370 - 392
outside
393 - 401
TMhelix
402 - 424
inside
425 - 436
TMhelix
437 - 459
outside
460 - 480
Population Genetic Test Statistics
Pi
21.534781
Theta
19.34258
Tajima's D
0.312466
CLR
0.038202
CSRT
0.462826858657067
Interpretation
Uncertain
