Gene
KWMTBOMO00273
Pre Gene Modal
BGIBMGA000550
Annotation
PREDICTED:_mediator_of_RNA_polymerase_II_transcription_subunit_23_[Amyelois_transitella]
Full name
Mediator of RNA polymerase II transcription subunit 23
Alternative Name
Mediator complex subunit 23
Location in the cell
PlasmaMembrane Reliability : 1.278
Sequence
CDS
ATGCCACTTTTTAGCAATGTTTTGCATTTAAAGGTACTACGTCCGAACCCAGATGGCAACGAAAGTCAAGTGTCACTCTACATCATTCAACTGTTGCTACTTCATGGTTTTGAGTTAAGAAACAGGGTTCAAGATTTCGTTAAGGAAAACTCACCAGAACACTGGAAACAAAATAATTGGTACGAAAAGCATTTAGCATTTCAAAGGAAATATCCCGAGAAGTTCACACCAGAAGAAGCGAGTGGTGCATATGGCGCACCCATACCCGTTTATTTGAGTAACGTTTGCTTAAGATTGATTCCTGTTTTTGATATAGTCGTACATAGACATCTGGAGATACATCAAGTTGCTAAATATCTAGAACAGCTATTGGAACATTTGGGATATCTGTATAAGTTTCATGATCGGCCTATAACCTTCCTTTACAATACTCTACACTATTACGAGAAGAAGCTACGCGATAAACCGTTGCTAAAGCGTAAACTGGTGGGAGCAGTATTGGGAGCACTCAAAGATGTCCGTCCCGCCGGCTGGGCCGCCACTGAAACGCTGCAAAATTTTCTGGCCAACACGGAGTATGATAACACTACGTGGGTTCCAGATCTTAATTATTATCTGAGTTTAGTCAATAGAATGGTGGACACAATGATGGGTAATTCTCACTTTCCAAATACTGACTGGAGATTCAACGAATACCCAAATCCGTCAGCACACGCCCTGTACGTAACTTGTGTCGAGTTGATGTCATTACCAGTTGCGCCTAATTACGTGGGGAACAGTTTACTCGATGTGATCACAAAAGGTTTCGTCGTAATACCCACCACGAAATTACAGCTTTGGATCAACGCTATTGGCTTAATAATGGCGGCTTTGCCGGACCCGTATTGGACGGTTCTGCACGAACGTCTTTTGGAACTTACGACTAGCAGTGAGATGATCGAATGGACATATCCATATTCACCTTTCCAGCTGTTTAATTTAGCAACCACAAGTGACGCGATGCTTGAGAACAAATATAGTTTGACATTGGCTTGCGCGCATGCCATATGGTACCATGCCGGCGCAGGGCAAATTATGCAAGTGCCTATTTTCGTAAAAGAGAAGCTGGCTGTTGAAATACACACGGAAACACAAATGCTCTTCTTGTGCCACATGGTGGGACCGTTCTTACAGAGATTTAATTCCGATCTACCGCGGACAGTGATGGACATAACTATAACTTTGTATGAACTACTCGCTCACATCGATAAGTCACAGACGCATCTTCAATATATGGATCCGATTTGCGACTTGCTATATCATATAAAATATATGTTCGTCGGCGACACATTGAAGAACGAAGTGGAAAACGTAATCCGTAAACTGCGACCGGCGCTGCAGATGCGTCTGCGCTTCATCACGCACTTGAATGTTGAACAAATTAACACCGCCTAG
Protein
MPLFSNVLHLKVLRPNPDGNESQVSLYIIQLLLLHGFELRNRVQDFVKENSPEHWKQNNWYEKHLAFQRKYPEKFTPEEASGAYGAPIPVYLSNVCLRLIPVFDIVVHRHLEIHQVAKYLEQLLEHLGYLYKFHDRPITFLYNTLHYYEKKLRDKPLLKRKLVGAVLGALKDVRPAGWAATETLQNFLANTEYDNTTWVPDLNYYLSLVNRMVDTMMGNSHFPNTDWRFNEYPNPSAHALYVTCVELMSLPVAPNYVGNSLLDVITKGFVVIPTTKLQLWINAIGLIMAALPDPYWTVLHERLLELTTSSEMIEWTYPYSPFQLFNLATTSDAMLENKYSLTLACAHAIWYHAGAGQIMQVPIFVKEKLAVEIHTETQMLFLCHMVGPFLQRFNSDLPRTVMDITITLYELLAHIDKSQTHLQYMDPICDLLYHIKYMFVGDTLKNEVENVIRKLRPALQMRLRFITHLNVEQINTA
Summary
Description
Component of the Mediator complex, a coactivator involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. Mediator functions as a bridge to convey information from gene-specific regulatory proteins to the basal RNA polymerase II transcription machinery. Mediator is recruited to promoters by direct interactions with regulatory proteins and serves as a scaffold for the assembly of a functional preinitiation complex with RNA polymerase II and the general transcription factors (By similarity).
Subunit
Component of the Mediator complex.
Similarity
Belongs to the Mediator complex subunit 23 family.
Keywords
Activator
Complete proteome
Nucleus
Reference proteome
Transcription
Transcription regulation
Feature
chain Mediator of RNA polymerase II transcription subunit 23
Uniprot
A0A2H1VRA5
A0A212EZE8
A0A194PRR3
A0A194QUV2
A0A2A4IRU5
H9ITH0
+ More
A0A067RQN7 A0A2J7PWY8 E0VW65 A0A1B6JFR5 A0A1B6I5N5 A0A1B6M771 A0A2A3E195 A0A0M8ZY05 A0A0L7R9A5 E2APD6 E9IBL3 A0A1B6D9V1 A0A1B6CFM9 A0A151WR11 A0A158NHZ3 F4WPV3 A0A195CKQ7 A0A154PJN6 A0A023F3N7 E2BEU2 A0A224XA35 A0A026WYL9 K7ISR1 A0A069DZS9 A0A195FDN0 A0A0C9QRJ8 A0A088AIA1 A0A195AWH8 A0A310S5F8 A0A291L1R5 A0A195EH54 A0A1B6HJQ1 A0A182QGM8 K7ISQ9 A0A0L7LV65 A0A182VM97 A0A182TUN9 A0A182NKF6 Q7PNT3 A0A232F082 A0A182MU65 A0A1Q3FB96 A0A182R728 A0A182WAZ8 A0A182PJT3 A0A182JBI5 A0A084W440 W5JH82 A0A2M4DQ68 A0A2M4AIU6 A0A182YR16 A0A182GVW2 B0VZI1 A0A0A9Z0N6 A0A146KUM8 A0A1Y1L9I3 A0A182XM38 Q16HH9 A0A182KPA8 A0A182I1M7 A0A182FD51 A0A182JX23 A0A1B0EZ22 A0A293MST9 A0A0P4VP90 A0A0P5A7M3 A0A0P5B6U5 A0A2H8TVI5 T1HR14 T1J7Z5 W8AP03 A0A0P5XVY6 D6WF99 A0A2S2QH62 A0A232F0E3 V5H511 V5H470 J9K0B7 A0A034V4Q1 A0A1B0G774 L7M0G0 A0A1E1X3C3 E9FU52 A0A0K8WAE7 A0A131YVN6 A0A224YV08 A0A2R5LDQ7 A0A0P5A583 A0A162EEV2 A0A0P5UPW5 A0A0P5LVD2 A0A0P5W921 A0A0P5RF41 A0A2S2NRG1
A0A067RQN7 A0A2J7PWY8 E0VW65 A0A1B6JFR5 A0A1B6I5N5 A0A1B6M771 A0A2A3E195 A0A0M8ZY05 A0A0L7R9A5 E2APD6 E9IBL3 A0A1B6D9V1 A0A1B6CFM9 A0A151WR11 A0A158NHZ3 F4WPV3 A0A195CKQ7 A0A154PJN6 A0A023F3N7 E2BEU2 A0A224XA35 A0A026WYL9 K7ISR1 A0A069DZS9 A0A195FDN0 A0A0C9QRJ8 A0A088AIA1 A0A195AWH8 A0A310S5F8 A0A291L1R5 A0A195EH54 A0A1B6HJQ1 A0A182QGM8 K7ISQ9 A0A0L7LV65 A0A182VM97 A0A182TUN9 A0A182NKF6 Q7PNT3 A0A232F082 A0A182MU65 A0A1Q3FB96 A0A182R728 A0A182WAZ8 A0A182PJT3 A0A182JBI5 A0A084W440 W5JH82 A0A2M4DQ68 A0A2M4AIU6 A0A182YR16 A0A182GVW2 B0VZI1 A0A0A9Z0N6 A0A146KUM8 A0A1Y1L9I3 A0A182XM38 Q16HH9 A0A182KPA8 A0A182I1M7 A0A182FD51 A0A182JX23 A0A1B0EZ22 A0A293MST9 A0A0P4VP90 A0A0P5A7M3 A0A0P5B6U5 A0A2H8TVI5 T1HR14 T1J7Z5 W8AP03 A0A0P5XVY6 D6WF99 A0A2S2QH62 A0A232F0E3 V5H511 V5H470 J9K0B7 A0A034V4Q1 A0A1B0G774 L7M0G0 A0A1E1X3C3 E9FU52 A0A0K8WAE7 A0A131YVN6 A0A224YV08 A0A2R5LDQ7 A0A0P5A583 A0A162EEV2 A0A0P5UPW5 A0A0P5LVD2 A0A0P5W921 A0A0P5RF41 A0A2S2NRG1
Pubmed
22118469
26354079
19121390
24845553
20566863
20798317
+ More
21282665 21347285 21719571 25474469 24508170 30249741 20075255 26334808 28964897 26227816 12364791 14747013 17210077 28648823 24438588 20920257 23761445 25244985 26483478 25401762 26823975 28004739 17510324 20966253 24495485 18362917 19820115 25765539 25348373 25576852 28503490 21292972 26830274 28797301
21282665 21347285 21719571 25474469 24508170 30249741 20075255 26334808 28964897 26227816 12364791 14747013 17210077 28648823 24438588 20920257 23761445 25244985 26483478 25401762 26823975 28004739 17510324 20966253 24495485 18362917 19820115 25765539 25348373 25576852 28503490 21292972 26830274 28797301
EMBL
ODYU01003957
SOQ43350.1
AGBW02011334
OWR46814.1
KQ459594
KPI96136.1
+ More
KQ461108 KPJ09247.1 NWSH01008035 PCG62717.1 BABH01006967 BABH01006968 BABH01006969 BABH01006970 BABH01006971 KK852480 KDR22950.1 NEVH01020867 PNF20839.1 DS235817 EEB17621.1 GECU01009702 JAS98004.1 GECU01025470 JAS82236.1 GEBQ01008207 JAT31770.1 KZ288462 PBC25467.1 KQ435828 KOX71879.1 KQ414627 KOC67460.1 GL441512 EFN64710.1 GL762111 EFZ22077.1 GEDC01014825 JAS22473.1 GEDC01025056 JAS12242.1 KQ982813 KYQ50339.1 ADTU01016114 ADTU01016115 GL888255 EGI63782.1 KQ977622 KYN01310.1 KQ434938 KZC12101.1 GBBI01002712 JAC16000.1 GL447857 EFN85808.1 GFTR01008652 JAW07774.1 KK107078 QOIP01000011 EZA60234.1 RLU17149.1 AAZX01002897 GBGD01000110 JAC88779.1 KQ981636 KYN38770.1 GBYB01006314 GBYB01006315 JAG76081.1 JAG76082.1 KQ976725 KYM76531.1 KQ769940 OAD52744.1 KY809857 ATI09806.1 KQ978957 KYN27194.1 GECU01032804 JAS74902.1 AXCN02000399 JTDY01000029 KOB79320.1 AAAB01008960 EAA11662.5 NNAY01001424 OXU24022.1 AXCM01004515 GFDL01010256 JAV24789.1 ATLV01020273 KE525297 KFB44984.1 ADMH02001390 ETN62683.1 GGFL01015110 MBW79288.1 GGFK01007394 MBW40715.1 JXUM01019778 JXUM01019779 KQ560554 KXJ81909.1 DS231814 EDS31718.1 GBHO01008229 JAG35375.1 GDHC01019314 JAP99314.1 GEZM01063768 JAV68970.1 CH478169 APCN01000069 AJVK01006587 GFWV01019215 MAA43943.1 GDRN01109245 JAI57099.1 GDIP01203910 JAJ19492.1 GDIP01203909 JAJ19493.1 GFXV01006442 MBW18247.1 ACPB03023227 ACPB03023228 JH431944 GAMC01018818 JAB87737.1 GDIP01066525 JAM37190.1 KQ971319 EFA00475.1 GGMS01007657 MBY76860.1 OXU24023.1 GANP01012409 JAB72059.1 GANP01006658 JAB77810.1 ABLF02029119 GAKP01020716 JAC38236.1 CCAG010008068 GACK01008475 JAA56559.1 GFAC01005450 JAT93738.1 GL732524 EFX89531.1 GDHF01004151 JAI48163.1 GEDV01005869 JAP82688.1 GFPF01009619 MAA20765.1 GGLE01003472 MBY07598.1 GDIP01203867 JAJ19535.1 LRGB01001937 KZS09960.1 GDIP01111276 JAL92438.1 GDIQ01164608 JAK87117.1 GDIP01089680 JAM14035.1 GDIQ01121113 JAL30613.1 GGMR01007116 MBY19735.1
KQ461108 KPJ09247.1 NWSH01008035 PCG62717.1 BABH01006967 BABH01006968 BABH01006969 BABH01006970 BABH01006971 KK852480 KDR22950.1 NEVH01020867 PNF20839.1 DS235817 EEB17621.1 GECU01009702 JAS98004.1 GECU01025470 JAS82236.1 GEBQ01008207 JAT31770.1 KZ288462 PBC25467.1 KQ435828 KOX71879.1 KQ414627 KOC67460.1 GL441512 EFN64710.1 GL762111 EFZ22077.1 GEDC01014825 JAS22473.1 GEDC01025056 JAS12242.1 KQ982813 KYQ50339.1 ADTU01016114 ADTU01016115 GL888255 EGI63782.1 KQ977622 KYN01310.1 KQ434938 KZC12101.1 GBBI01002712 JAC16000.1 GL447857 EFN85808.1 GFTR01008652 JAW07774.1 KK107078 QOIP01000011 EZA60234.1 RLU17149.1 AAZX01002897 GBGD01000110 JAC88779.1 KQ981636 KYN38770.1 GBYB01006314 GBYB01006315 JAG76081.1 JAG76082.1 KQ976725 KYM76531.1 KQ769940 OAD52744.1 KY809857 ATI09806.1 KQ978957 KYN27194.1 GECU01032804 JAS74902.1 AXCN02000399 JTDY01000029 KOB79320.1 AAAB01008960 EAA11662.5 NNAY01001424 OXU24022.1 AXCM01004515 GFDL01010256 JAV24789.1 ATLV01020273 KE525297 KFB44984.1 ADMH02001390 ETN62683.1 GGFL01015110 MBW79288.1 GGFK01007394 MBW40715.1 JXUM01019778 JXUM01019779 KQ560554 KXJ81909.1 DS231814 EDS31718.1 GBHO01008229 JAG35375.1 GDHC01019314 JAP99314.1 GEZM01063768 JAV68970.1 CH478169 APCN01000069 AJVK01006587 GFWV01019215 MAA43943.1 GDRN01109245 JAI57099.1 GDIP01203910 JAJ19492.1 GDIP01203909 JAJ19493.1 GFXV01006442 MBW18247.1 ACPB03023227 ACPB03023228 JH431944 GAMC01018818 JAB87737.1 GDIP01066525 JAM37190.1 KQ971319 EFA00475.1 GGMS01007657 MBY76860.1 OXU24023.1 GANP01012409 JAB72059.1 GANP01006658 JAB77810.1 ABLF02029119 GAKP01020716 JAC38236.1 CCAG010008068 GACK01008475 JAA56559.1 GFAC01005450 JAT93738.1 GL732524 EFX89531.1 GDHF01004151 JAI48163.1 GEDV01005869 JAP82688.1 GFPF01009619 MAA20765.1 GGLE01003472 MBY07598.1 GDIP01203867 JAJ19535.1 LRGB01001937 KZS09960.1 GDIP01111276 JAL92438.1 GDIQ01164608 JAK87117.1 GDIP01089680 JAM14035.1 GDIQ01121113 JAL30613.1 GGMR01007116 MBY19735.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000005204
UP000027135
+ More
UP000235965 UP000009046 UP000242457 UP000053105 UP000053825 UP000000311 UP000075809 UP000005205 UP000007755 UP000078542 UP000076502 UP000008237 UP000053097 UP000279307 UP000002358 UP000078541 UP000005203 UP000078540 UP000078492 UP000075886 UP000037510 UP000075903 UP000075902 UP000075884 UP000007062 UP000215335 UP000075883 UP000075900 UP000075920 UP000075885 UP000075880 UP000030765 UP000000673 UP000076408 UP000069940 UP000249989 UP000002320 UP000076407 UP000008820 UP000075882 UP000075840 UP000069272 UP000075881 UP000092462 UP000015103 UP000007266 UP000007819 UP000092444 UP000000305 UP000076858
UP000235965 UP000009046 UP000242457 UP000053105 UP000053825 UP000000311 UP000075809 UP000005205 UP000007755 UP000078542 UP000076502 UP000008237 UP000053097 UP000279307 UP000002358 UP000078541 UP000005203 UP000078540 UP000078492 UP000075886 UP000037510 UP000075903 UP000075902 UP000075884 UP000007062 UP000215335 UP000075883 UP000075900 UP000075920 UP000075885 UP000075880 UP000030765 UP000000673 UP000076408 UP000069940 UP000249989 UP000002320 UP000076407 UP000008820 UP000075882 UP000075840 UP000069272 UP000075881 UP000092462 UP000015103 UP000007266 UP000007819 UP000092444 UP000000305 UP000076858
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
A0A2H1VRA5
A0A212EZE8
A0A194PRR3
A0A194QUV2
A0A2A4IRU5
H9ITH0
+ More
A0A067RQN7 A0A2J7PWY8 E0VW65 A0A1B6JFR5 A0A1B6I5N5 A0A1B6M771 A0A2A3E195 A0A0M8ZY05 A0A0L7R9A5 E2APD6 E9IBL3 A0A1B6D9V1 A0A1B6CFM9 A0A151WR11 A0A158NHZ3 F4WPV3 A0A195CKQ7 A0A154PJN6 A0A023F3N7 E2BEU2 A0A224XA35 A0A026WYL9 K7ISR1 A0A069DZS9 A0A195FDN0 A0A0C9QRJ8 A0A088AIA1 A0A195AWH8 A0A310S5F8 A0A291L1R5 A0A195EH54 A0A1B6HJQ1 A0A182QGM8 K7ISQ9 A0A0L7LV65 A0A182VM97 A0A182TUN9 A0A182NKF6 Q7PNT3 A0A232F082 A0A182MU65 A0A1Q3FB96 A0A182R728 A0A182WAZ8 A0A182PJT3 A0A182JBI5 A0A084W440 W5JH82 A0A2M4DQ68 A0A2M4AIU6 A0A182YR16 A0A182GVW2 B0VZI1 A0A0A9Z0N6 A0A146KUM8 A0A1Y1L9I3 A0A182XM38 Q16HH9 A0A182KPA8 A0A182I1M7 A0A182FD51 A0A182JX23 A0A1B0EZ22 A0A293MST9 A0A0P4VP90 A0A0P5A7M3 A0A0P5B6U5 A0A2H8TVI5 T1HR14 T1J7Z5 W8AP03 A0A0P5XVY6 D6WF99 A0A2S2QH62 A0A232F0E3 V5H511 V5H470 J9K0B7 A0A034V4Q1 A0A1B0G774 L7M0G0 A0A1E1X3C3 E9FU52 A0A0K8WAE7 A0A131YVN6 A0A224YV08 A0A2R5LDQ7 A0A0P5A583 A0A162EEV2 A0A0P5UPW5 A0A0P5LVD2 A0A0P5W921 A0A0P5RF41 A0A2S2NRG1
A0A067RQN7 A0A2J7PWY8 E0VW65 A0A1B6JFR5 A0A1B6I5N5 A0A1B6M771 A0A2A3E195 A0A0M8ZY05 A0A0L7R9A5 E2APD6 E9IBL3 A0A1B6D9V1 A0A1B6CFM9 A0A151WR11 A0A158NHZ3 F4WPV3 A0A195CKQ7 A0A154PJN6 A0A023F3N7 E2BEU2 A0A224XA35 A0A026WYL9 K7ISR1 A0A069DZS9 A0A195FDN0 A0A0C9QRJ8 A0A088AIA1 A0A195AWH8 A0A310S5F8 A0A291L1R5 A0A195EH54 A0A1B6HJQ1 A0A182QGM8 K7ISQ9 A0A0L7LV65 A0A182VM97 A0A182TUN9 A0A182NKF6 Q7PNT3 A0A232F082 A0A182MU65 A0A1Q3FB96 A0A182R728 A0A182WAZ8 A0A182PJT3 A0A182JBI5 A0A084W440 W5JH82 A0A2M4DQ68 A0A2M4AIU6 A0A182YR16 A0A182GVW2 B0VZI1 A0A0A9Z0N6 A0A146KUM8 A0A1Y1L9I3 A0A182XM38 Q16HH9 A0A182KPA8 A0A182I1M7 A0A182FD51 A0A182JX23 A0A1B0EZ22 A0A293MST9 A0A0P4VP90 A0A0P5A7M3 A0A0P5B6U5 A0A2H8TVI5 T1HR14 T1J7Z5 W8AP03 A0A0P5XVY6 D6WF99 A0A2S2QH62 A0A232F0E3 V5H511 V5H470 J9K0B7 A0A034V4Q1 A0A1B0G774 L7M0G0 A0A1E1X3C3 E9FU52 A0A0K8WAE7 A0A131YVN6 A0A224YV08 A0A2R5LDQ7 A0A0P5A583 A0A162EEV2 A0A0P5UPW5 A0A0P5LVD2 A0A0P5W921 A0A0P5RF41 A0A2S2NRG1
PDB
6H02
E-value=1.51049e-136,
Score=1246
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
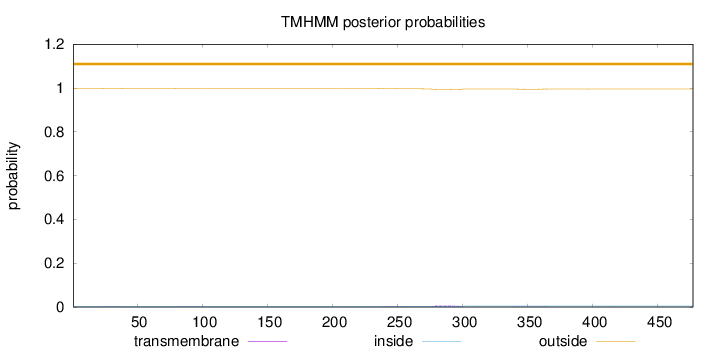
Length:
477
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.23517
Exp number, first 60 AAs:
0.00544
Total prob of N-in:
0.00106
outside
1 - 477
Population Genetic Test Statistics
Pi
19.166848
Theta
27.805772
Tajima's D
-1.340117
CLR
0.008334
CSRT
0.0787460626968652
Interpretation
Uncertain
