Pre Gene Modal
BGIBMGA000553
Annotation
PREDICTED:_peroxidasin_isoform_X2_[Bombyx_mori]
Full name
Peroxidasin
Location in the cell
Cytoplasmic Reliability : 1.663 Nuclear Reliability : 2.085
Sequence
CDS
ATGCCAAAGTTAAGGAAACTCCGTTTAGATAGTAATGCATTGATCTGCGATTGTTCTATACTATGGTTCATCCGTATGCTGGAGAATAACGAATGGATGCATGTGGCGGCAACTTGTTACCAACCAGCCTCCGTCACTGGAACCTCCCTAGCCGCTATGAAGCACCATGACCTTCCTTGCCAACAACCTCAATTCGAATCCGAACCAAACGATGTTGAAGTGAGTTTTGGAGAAAATGCAGTTTTCACATGTGTGGCGACTGGTGAACCAGCACCTGAAATTCTTTGGCTTCGTGATTCAGAAGTTATTCCATTAAATGAAAATCGATACGAAATTTTCGACAACGGAACGCTTATGGTTCATGAAGCTAATGAAAACGATGATGGCTTGTTTGAATGCATGGCAAAAAATGTGGCAGGAAAAGCTCATTCTAAACCGGCAAAAATGATTGTACGAAGAAAAGCGGAAGAACATAAAAGTCCCGCTCCTCCGGTATTCGTTTCGACGCCACAAAAACGATTCGCCAAAATTAATGAAACTTCGGTGATGTTTGATTGCACTGCTACTGGATTTCCCAAGCCGCATGTGGCCTGGTATTTTAACGGAGAAAGAATGTTATTAAACGAGCGCATAATTTTACAACATAATGGATCTATTCTTATCAAAAATGTTCAAAATTCAGATGCGGGTGTTTACACATGTCAAGTTGAAAACATACATGGAAAAATATCTGCATCTGTTCCATTAGAGGTTACTGCTCCGCCATCTTTTATAGTGGTTCCCACGAATCAGACAGCTATTATCGGAGATAACGTTTGGTTTACGTGTAAAGCGAAGGGAACTCCAAAACCTTCGATTAAATGGTATAGAAATACGGTAATATTACCTATAACTGAAAATATTGTGCTGAGTGATGATAATCAAAATTTAACTTTATTGGAAGTGACTAGAGATGATGATGCTATATATCACTGCAGAGCAGAAAATGATGGAGGTATGATTGAAGCATCTGCTGTTCTAACACTCGAACAGGTAGAAATCATTCCTCCTCGTATCGTTTTGAAGCCAGAAGATATGGATGCATTTAAAGAAACTTCAGTGCAAATGCCTTGCGAAATCGAAAGTGTGCCACCTGCTTCGATTGATTGGAGGAAAGATGGTTCAAGAATCGTAGAAGATGATAGAATTTCGATAACAGTGATTGGTAGCCTCATAATAAGAAACGTCTCCGTAACCGATACTGGTCGATATGAATGTTCAGCTTTTAATGATTATGGTCGTGATACAGCATCCGTTTTTCTTACTGTTAAGGACAACCTTTTACCAGGGGACGCGTACGTAAACATAGCTTTGACACAAGCTATGCGGGACGTAGACCAAGCCATTGAAAGAACAGTTGACCAATTATTTAAAAATCGAAGTGCCGTAGTTAATATCGGCGATCTTTATAGAATGACCAGGTTCCCTAATGCACCCGAACGTGAAGTAGCAAGAGCAGCAGAAATTTATGAAAGGACTTTAGATAAATTAAAAGAATATATACAGAGCGGAAAGAAGTTAAGTACAGCTGAACCATTTAATTATCAAAGCGTACTATCAAACCAACACCTCGAAGTAATTGCAAGACTATCTGGATGTGTGGCCCACAGAGAAGAAAAAGACTGCTCTGACATGTGTTACCATAGTAAATACCGTTCACTTGATGGCAGTTGTAACAACTTTGCACATCCTATGTGGGGTACATCACTTTCAGGATTCAGAAGAATTCTTTATCCTATTTACGAAAACGGTTTTAGCCAACCAATTGGGTGGAATAAAGATTTAAAATATAACGGTTTTACATTACCAAGTGCACGATTGATATCTACTGCTATTATAACGACTACCGAAATAACACAGGATATAGAAATAACGCATATGACTATGCAATGGGGACAGTGGTTAGATCATGATTTGGATCACGCATTACCCCCTGTAAGTTCTCAAACTTGGGATGGTGTTGATTGTAAGAAAACTTGCGACTATGCTCCTCCTTGTTTTCCGATTGATGTACCCTTAAACGATCCTAGAGTGAATAATCGGCGATGCATTGATTTTATAAGAACGAGCTCTGTTTGTGGTTCAGGAACAACTTCTGTTCTTTTCGGTACTTTGCAAACAAGAGAACAGATTAATCAACTTACTTCTTTTCTTGACGCTTCACAAGTGTACGGTTTTGAAAAATCAGTGGCAGAAGACCTTCGTGATCTAACCAATGATAATGGATTACTCAGAGTCGGTGCCACATTTCCTGGTAGAAAACCGTTACTACCAACATCAGGCTTAAACGGAATGGACTGTCGACGTAATTTAGAAGAGAGCACGCGAAGTTGTTTTGTGGCTGGTGATATTAGAGCAAATGAGCAAATTGGTCTAGCAGCTATGCACACCATTTGGATGCGAGAGCACAATCGTATAGCCCTTCAATTAAAAGCCTTGAATCCATTTTGGGATGGCGACAGAGTTTACCAAGAAGCAAGAAAAGTTGTGGGTGCTCAAATGCAATTTATCACTTATGAACATTGGCTGCCTATCATATTAGGTCCGGAAGGATATGATCAATTGGGAAAATATAAACAATACGATAATCGTCTTAATCCATCAATATCTAATGTTTTTGCCACAGCTGCTCTTCGCTTTGGACATTCTTTGATCAATCCTGTACTTCATCGGTATGACGAAAACTTTGAAACTATACCCCAGGGTCACTTATTGCTTCGTAATGCATTTTTTTCACCATGGAGATTAGTTGACGAAGGTGGAGTAGACCCTTTGCTGCGAGGAATGTTTACAACACCAGCAAAATTGAAAACATCTAAACAAAATCTTAATTCAGAACTGACGGAAAAGCTATTCTACAGCGCACATGCTGTAGCTCTTGATTTAGCTGCCATTAATATACAAAGGGGCCGAGACCACGCTATACCTCCATACACTAAGTGGCGTCAGTTTTGTAATATGAGTGAAGTTAACGACTTTGACGATTTGTCTGGTGAAATAAGCGATAAAGAAGTAAGGGACAAATTACAGGAGCTTTACGGATCAGTTCACAATATAGATGTTTGGGTAGGAGGAATTTTAGAAGATCAGGTTGAAGGTGGCAAAGTCGGTCCCTTATTTAGATGTTTATTAATGGAGCAATTTGTTCGTTTACGCGATGGCGATCGATTTTGGTATGAAAACCCAAGCGTGTTTAAACCCGATCAACTTCGACAGATAAAGGAAACAAGTCTAGCTCGAATCTTGTGTGACAATGGCGATAATATTGATACAATTAGCGAAAACGTATTTTACTTACCGGAAGTACAAGATGGGCTTGTATCTTGTGAAGATTTGCCATCAATGGATCTTCGTTTCTGGGCTGACTGTGAATCTTGTAATTTTGATCTTGATACAGAGAGAGAGAAGCGGCGAGCTCGAAGAGATGTTGATTCCAATATTACAAACGGCCGCTTTAATAAATTGGAACATTTACAAAATGATTTAGTACATACCATTGAGATGCTTAAGAAAAGAATCGAAATACTCGAAGCTACCTGTAAAGCGAACGATACTCTTTCTTTCAGTAATGAATTTCCATTCGAGGAAGAGTAA
Protein
MPKLRKLRLDSNALICDCSILWFIRMLENNEWMHVAATCYQPASVTGTSLAAMKHHDLPCQQPQFESEPNDVEVSFGENAVFTCVATGEPAPEILWLRDSEVIPLNENRYEIFDNGTLMVHEANENDDGLFECMAKNVAGKAHSKPAKMIVRRKAEEHKSPAPPVFVSTPQKRFAKINETSVMFDCTATGFPKPHVAWYFNGERMLLNERIILQHNGSILIKNVQNSDAGVYTCQVENIHGKISASVPLEVTAPPSFIVVPTNQTAIIGDNVWFTCKAKGTPKPSIKWYRNTVILPITENIVLSDDNQNLTLLEVTRDDDAIYHCRAENDGGMIEASAVLTLEQVEIIPPRIVLKPEDMDAFKETSVQMPCEIESVPPASIDWRKDGSRIVEDDRISITVIGSLIIRNVSVTDTGRYECSAFNDYGRDTASVFLTVKDNLLPGDAYVNIALTQAMRDVDQAIERTVDQLFKNRSAVVNIGDLYRMTRFPNAPEREVARAAEIYERTLDKLKEYIQSGKKLSTAEPFNYQSVLSNQHLEVIARLSGCVAHREEKDCSDMCYHSKYRSLDGSCNNFAHPMWGTSLSGFRRILYPIYENGFSQPIGWNKDLKYNGFTLPSARLISTAIITTTEITQDIEITHMTMQWGQWLDHDLDHALPPVSSQTWDGVDCKKTCDYAPPCFPIDVPLNDPRVNNRRCIDFIRTSSVCGSGTTSVLFGTLQTREQINQLTSFLDASQVYGFEKSVAEDLRDLTNDNGLLRVGATFPGRKPLLPTSGLNGMDCRRNLEESTRSCFVAGDIRANEQIGLAAMHTIWMREHNRIALQLKALNPFWDGDRVYQEARKVVGAQMQFITYEHWLPIILGPEGYDQLGKYKQYDNRLNPSISNVFATAALRFGHSLINPVLHRYDENFETIPQGHLLLRNAFFSPWRLVDEGGVDPLLRGMFTTPAKLKTSKQNLNSELTEKLFYSAHAVALDLAAINIQRGRDHAIPPYTKWRQFCNMSEVNDFDDLSGEISDKEVRDKLQELYGSVHNIDVWVGGILEDQVEGGKVGPLFRCLLMEQFVRLRDGDRFWYENPSVFKPDQLRQIKETSLARILCDNGDNIDTISENVFYLPEVQDGLVSCEDLPSMDLRFWADCESCNFDLDTEREKRRARRDVDSNITNGRFNKLEHLQNDLVHTIEMLKKRIEILEATCKANDTLSFSNEFPFEEE
Summary
Description
Plays a role in extracellular matrix consolidation, phagocytosis and defense.
Catalytic Activity
2 a phenolic donor + H2O2 = 2 a phenolic radical donor + 2 H2O
Cofactor
Ca(2+)
heme b
heme b
Subunit
Homotrimer; disulfide-linked.
Similarity
Belongs to the peroxidase family. XPO subfamily.
Keywords
Alternative splicing
Calcium
Coiled coil
Complete proteome
Disulfide bond
Glycoprotein
Heme
Hydrogen peroxide
Immunoglobulin domain
Iron
Leucine-rich repeat
Metal-binding
Oxidoreductase
Peroxidase
Reference proteome
Repeat
Secreted
Signal
Feature
chain Peroxidasin
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9ITH3
A0A2H1WBJ7
A0A2W1BU53
A0A212EZ95
A0A2A4K172
A0A194QV98
+ More
A0A194PYB4 A0A0L7LV38 A0A0K0Q0G3 D6WFS4 A0A1Y1KP07 A0A1W4XC42 N6U3L9 V5GJ36 A0A0T6BC77 Q7QJ29 A0A1B6E548 A0A1B6EDT6 A0A1B6DAD0 A0A1B6CFM4 A0A1B6DJE2 A0A1J1IUS8 A0A084VQX4 Q2UZU6 A0A1B6K977 B4KYM2 A0A182UXG5 A0A182QQU6 A0A2C9GPS4 A0A2P8Y7Y4 B4N392 W5JXD0 A0A0M3QW13 D2NUH5 Q9VZZ4 B4H1J9 Q29FB4 E0R979 A0A182KQP7 B3NBK0 B4IZC8 A0A0J9RML2 B4QNK1 A0A3B0KKT0 A0A1W4VP18 A0A1S4G837 E0VZU0 B3M7Y3 A0A2S2QJH4 B4PDD3 A0A2S2NP02 A0A2H8TLB2 A0A0L0C335 B4LDU2 A0A1I8MZ49 T1PGN3 A0A1I8QDU7 A0A0N7ZAC2 A0A0P4VSH7 A0A2P2HZE6 T1DFR8 A0A0P4VX44 A0A0P6IDE8 A0A0P6DAX5 A0A0P5ZY75 A0A0N8EFR5 A0A0P6GUX0 A0A0P5GP60 A0A0P5M4M6 A0A0N7ZKD4 A0A2A3EPZ0 A0A088A0S4 A0A0P6BZV6 Q17PE4 A0A034VZ39 A0A2L2YGD2 A0A2L2YGP6 A0A1A9V3P3 A0A2S2NNG2 A0A0C9QK01 A0A0C9Q881 A0A087U8G0 W8B783 A0A182TDB8 A0A0A1XJI6 A0A0N0BCT9 A0A182MJS5 K7ISQ6 A0A182WJQ7 A0A151XHN3 A0A182XLK9 A0A182PTY0 A0A182R3I0 A0A182JPM6 A0A0L7QX38 A0A1B0G8G3 A0A310SEP7 A0A232F4G8 A0A3B0KFL9 A0A0J7L8E5
A0A194PYB4 A0A0L7LV38 A0A0K0Q0G3 D6WFS4 A0A1Y1KP07 A0A1W4XC42 N6U3L9 V5GJ36 A0A0T6BC77 Q7QJ29 A0A1B6E548 A0A1B6EDT6 A0A1B6DAD0 A0A1B6CFM4 A0A1B6DJE2 A0A1J1IUS8 A0A084VQX4 Q2UZU6 A0A1B6K977 B4KYM2 A0A182UXG5 A0A182QQU6 A0A2C9GPS4 A0A2P8Y7Y4 B4N392 W5JXD0 A0A0M3QW13 D2NUH5 Q9VZZ4 B4H1J9 Q29FB4 E0R979 A0A182KQP7 B3NBK0 B4IZC8 A0A0J9RML2 B4QNK1 A0A3B0KKT0 A0A1W4VP18 A0A1S4G837 E0VZU0 B3M7Y3 A0A2S2QJH4 B4PDD3 A0A2S2NP02 A0A2H8TLB2 A0A0L0C335 B4LDU2 A0A1I8MZ49 T1PGN3 A0A1I8QDU7 A0A0N7ZAC2 A0A0P4VSH7 A0A2P2HZE6 T1DFR8 A0A0P4VX44 A0A0P6IDE8 A0A0P6DAX5 A0A0P5ZY75 A0A0N8EFR5 A0A0P6GUX0 A0A0P5GP60 A0A0P5M4M6 A0A0N7ZKD4 A0A2A3EPZ0 A0A088A0S4 A0A0P6BZV6 Q17PE4 A0A034VZ39 A0A2L2YGD2 A0A2L2YGP6 A0A1A9V3P3 A0A2S2NNG2 A0A0C9QK01 A0A0C9Q881 A0A087U8G0 W8B783 A0A182TDB8 A0A0A1XJI6 A0A0N0BCT9 A0A182MJS5 K7ISQ6 A0A182WJQ7 A0A151XHN3 A0A182XLK9 A0A182PTY0 A0A182R3I0 A0A182JPM6 A0A0L7QX38 A0A1B0G8G3 A0A310SEP7 A0A232F4G8 A0A3B0KFL9 A0A0J7L8E5
EC Number
1.11.1.7
Pubmed
19121390
28756777
22118469
26354079
26227816
18362917
+ More
19820115 28004739 23537049 12364791 24438588 17994087 29403074 20920257 23761445 8062820 10731132 12537572 12537569 17893096 15632085 20966253 22936249 17510324 20566863 17550304 26108605 25315136 25348373 26561354 24495485 25830018 20075255 28648823
19820115 28004739 23537049 12364791 24438588 17994087 29403074 20920257 23761445 8062820 10731132 12537572 12537569 17893096 15632085 20966253 22936249 17510324 20566863 17550304 26108605 25315136 25348373 26561354 24495485 25830018 20075255 28648823
EMBL
BABH01006966
ODYU01007557
SOQ50430.1
KZ149961
PZC76310.1
AGBW02011334
+ More
OWR46812.1 NWSH01000249 PCG78021.1 KQ461108 KPJ09244.1 KQ459594 KPI96140.1 JTDY01000029 KOB79317.1 KP727757 AKR04374.1 KQ971319 EFA01243.1 GEZM01077924 JAV63142.1 APGK01050877 APGK01050878 APGK01050879 KB741178 ENN73167.1 GALX01004387 JAB64079.1 LJIG01002437 KRT84503.1 AAAB01008807 EAA04206.4 GEDC01004242 JAS33056.1 GEDC01012000 GEDC01003931 GEDC01001217 JAS25298.1 JAS33367.1 JAS36081.1 GEDC01030471 GEDC01030383 GEDC01014753 JAS06827.1 JAS06915.1 JAS22545.1 GEDC01025070 JAS12228.1 GEDC01011477 JAS25821.1 CVRI01000059 CRL03332.1 ATLV01015338 ATLV01015339 ATLV01015340 KE525006 KFB40368.1 AF484235 AAQ05970.1 GEBQ01032002 JAT07975.1 CH933809 EDW17736.2 AXCN02000541 APCN01000179 PYGN01000819 PSN40372.1 CH964062 EDW78831.1 ADMH02000008 ETN68124.1 CP012525 ALC43349.1 BT120118 ADB25334.1 U11052 AE014296 AY051536 AY052120 CH479202 EDW30176.1 CH379067 EAL31341.2 BT125647 ADN05224.1 CH954178 EDV50322.1 CH916366 EDV96683.1 CM002912 KMY97161.1 KMY97162.1 CM000363 EDX08964.1 OUUW01000012 SPP87149.1 DS235854 EEB18896.1 CH902618 EDV39891.2 GGMS01008684 MBY77887.1 CM000159 EDW93913.1 KRK01498.1 GGMR01006063 MBY18682.1 GFXV01002657 MBW14462.1 JRES01000966 KNC26676.1 CH940647 EDW68965.2 KA647859 AFP62488.1 GDRN01101085 JAI58440.1 GDRN01101086 JAI58439.1 IACF01001451 LAB67151.1 GAKT01000146 JAA92916.1 GDRN01101081 JAI58442.1 GDIQ01031847 JAN62890.1 GDIQ01079925 JAN14812.1 GDIP01050331 JAM53384.1 GDIQ01031148 JAN63589.1 GDIQ01028905 JAN65832.1 GDIQ01239780 JAK11945.1 GDIQ01169447 JAK82278.1 GDIP01236865 JAI86536.1 KZ288201 PBC33574.1 GDIP01011716 JAM91999.1 CH477191 EAT48600.2 GAKP01011595 JAC47357.1 IAAA01025214 LAA07216.1 IAAA01025215 IAAA01025216 LAA07217.1 GGMR01006104 MBY18723.1 GBYB01015138 JAG84905.1 GBYB01010428 JAG80195.1 KK118706 KFM73649.1 GAMC01012118 JAB94437.1 GBXI01003609 JAD10683.1 KQ435891 KOX69438.1 AXCM01002371 AAZX01002897 KQ982130 KYQ59821.1 KQ414705 KOC63131.1 CCAG010000251 KQ767283 OAD53466.1 NNAY01001047 OXU25353.1 SPP87150.1 LBMM01000218 KMR04342.1
OWR46812.1 NWSH01000249 PCG78021.1 KQ461108 KPJ09244.1 KQ459594 KPI96140.1 JTDY01000029 KOB79317.1 KP727757 AKR04374.1 KQ971319 EFA01243.1 GEZM01077924 JAV63142.1 APGK01050877 APGK01050878 APGK01050879 KB741178 ENN73167.1 GALX01004387 JAB64079.1 LJIG01002437 KRT84503.1 AAAB01008807 EAA04206.4 GEDC01004242 JAS33056.1 GEDC01012000 GEDC01003931 GEDC01001217 JAS25298.1 JAS33367.1 JAS36081.1 GEDC01030471 GEDC01030383 GEDC01014753 JAS06827.1 JAS06915.1 JAS22545.1 GEDC01025070 JAS12228.1 GEDC01011477 JAS25821.1 CVRI01000059 CRL03332.1 ATLV01015338 ATLV01015339 ATLV01015340 KE525006 KFB40368.1 AF484235 AAQ05970.1 GEBQ01032002 JAT07975.1 CH933809 EDW17736.2 AXCN02000541 APCN01000179 PYGN01000819 PSN40372.1 CH964062 EDW78831.1 ADMH02000008 ETN68124.1 CP012525 ALC43349.1 BT120118 ADB25334.1 U11052 AE014296 AY051536 AY052120 CH479202 EDW30176.1 CH379067 EAL31341.2 BT125647 ADN05224.1 CH954178 EDV50322.1 CH916366 EDV96683.1 CM002912 KMY97161.1 KMY97162.1 CM000363 EDX08964.1 OUUW01000012 SPP87149.1 DS235854 EEB18896.1 CH902618 EDV39891.2 GGMS01008684 MBY77887.1 CM000159 EDW93913.1 KRK01498.1 GGMR01006063 MBY18682.1 GFXV01002657 MBW14462.1 JRES01000966 KNC26676.1 CH940647 EDW68965.2 KA647859 AFP62488.1 GDRN01101085 JAI58440.1 GDRN01101086 JAI58439.1 IACF01001451 LAB67151.1 GAKT01000146 JAA92916.1 GDRN01101081 JAI58442.1 GDIQ01031847 JAN62890.1 GDIQ01079925 JAN14812.1 GDIP01050331 JAM53384.1 GDIQ01031148 JAN63589.1 GDIQ01028905 JAN65832.1 GDIQ01239780 JAK11945.1 GDIQ01169447 JAK82278.1 GDIP01236865 JAI86536.1 KZ288201 PBC33574.1 GDIP01011716 JAM91999.1 CH477191 EAT48600.2 GAKP01011595 JAC47357.1 IAAA01025214 LAA07216.1 IAAA01025215 IAAA01025216 LAA07217.1 GGMR01006104 MBY18723.1 GBYB01015138 JAG84905.1 GBYB01010428 JAG80195.1 KK118706 KFM73649.1 GAMC01012118 JAB94437.1 GBXI01003609 JAD10683.1 KQ435891 KOX69438.1 AXCM01002371 AAZX01002897 KQ982130 KYQ59821.1 KQ414705 KOC63131.1 CCAG010000251 KQ767283 OAD53466.1 NNAY01001047 OXU25353.1 SPP87150.1 LBMM01000218 KMR04342.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000037510
+ More
UP000007266 UP000192223 UP000019118 UP000007062 UP000183832 UP000030765 UP000009192 UP000075903 UP000075886 UP000075840 UP000245037 UP000007798 UP000000673 UP000092553 UP000000803 UP000008744 UP000001819 UP000075882 UP000008711 UP000001070 UP000000304 UP000268350 UP000192221 UP000009046 UP000007801 UP000002282 UP000037069 UP000008792 UP000095301 UP000095300 UP000242457 UP000005203 UP000008820 UP000078200 UP000054359 UP000075902 UP000053105 UP000075883 UP000002358 UP000075920 UP000075809 UP000076407 UP000075885 UP000075900 UP000075881 UP000053825 UP000092444 UP000215335 UP000036403
UP000007266 UP000192223 UP000019118 UP000007062 UP000183832 UP000030765 UP000009192 UP000075903 UP000075886 UP000075840 UP000245037 UP000007798 UP000000673 UP000092553 UP000000803 UP000008744 UP000001819 UP000075882 UP000008711 UP000001070 UP000000304 UP000268350 UP000192221 UP000009046 UP000007801 UP000002282 UP000037069 UP000008792 UP000095301 UP000095300 UP000242457 UP000005203 UP000008820 UP000078200 UP000054359 UP000075902 UP000053105 UP000075883 UP000002358 UP000075920 UP000075809 UP000076407 UP000075885 UP000075900 UP000075881 UP000053825 UP000092444 UP000215335 UP000036403
Pfam
Interpro
IPR007110
Ig-like_dom
+ More
IPR037120 Haem_peroxidase_sf_animal
IPR003599 Ig_sub
IPR019791 Haem_peroxidase_animal
IPR000483 Cys-rich_flank_reg_C
IPR013783 Ig-like_fold
IPR032675 LRR_dom_sf
IPR013098 Ig_I-set
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR034824 Peroxidasin_peroxidase
IPR003598 Ig_sub2
IPR034828 Peroxidasin
IPR036179 Ig-like_dom_sf
IPR010255 Haem_peroxidase_sf
IPR013106 Ig_V-set
IPR001007 VWF_dom
IPR000372 LRRNT
IPR026906 LRR_5
IPR037120 Haem_peroxidase_sf_animal
IPR003599 Ig_sub
IPR019791 Haem_peroxidase_animal
IPR000483 Cys-rich_flank_reg_C
IPR013783 Ig-like_fold
IPR032675 LRR_dom_sf
IPR013098 Ig_I-set
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR034824 Peroxidasin_peroxidase
IPR003598 Ig_sub2
IPR034828 Peroxidasin
IPR036179 Ig-like_dom_sf
IPR010255 Haem_peroxidase_sf
IPR013106 Ig_V-set
IPR001007 VWF_dom
IPR000372 LRRNT
IPR026906 LRR_5
Gene 3D
ProteinModelPortal
H9ITH3
A0A2H1WBJ7
A0A2W1BU53
A0A212EZ95
A0A2A4K172
A0A194QV98
+ More
A0A194PYB4 A0A0L7LV38 A0A0K0Q0G3 D6WFS4 A0A1Y1KP07 A0A1W4XC42 N6U3L9 V5GJ36 A0A0T6BC77 Q7QJ29 A0A1B6E548 A0A1B6EDT6 A0A1B6DAD0 A0A1B6CFM4 A0A1B6DJE2 A0A1J1IUS8 A0A084VQX4 Q2UZU6 A0A1B6K977 B4KYM2 A0A182UXG5 A0A182QQU6 A0A2C9GPS4 A0A2P8Y7Y4 B4N392 W5JXD0 A0A0M3QW13 D2NUH5 Q9VZZ4 B4H1J9 Q29FB4 E0R979 A0A182KQP7 B3NBK0 B4IZC8 A0A0J9RML2 B4QNK1 A0A3B0KKT0 A0A1W4VP18 A0A1S4G837 E0VZU0 B3M7Y3 A0A2S2QJH4 B4PDD3 A0A2S2NP02 A0A2H8TLB2 A0A0L0C335 B4LDU2 A0A1I8MZ49 T1PGN3 A0A1I8QDU7 A0A0N7ZAC2 A0A0P4VSH7 A0A2P2HZE6 T1DFR8 A0A0P4VX44 A0A0P6IDE8 A0A0P6DAX5 A0A0P5ZY75 A0A0N8EFR5 A0A0P6GUX0 A0A0P5GP60 A0A0P5M4M6 A0A0N7ZKD4 A0A2A3EPZ0 A0A088A0S4 A0A0P6BZV6 Q17PE4 A0A034VZ39 A0A2L2YGD2 A0A2L2YGP6 A0A1A9V3P3 A0A2S2NNG2 A0A0C9QK01 A0A0C9Q881 A0A087U8G0 W8B783 A0A182TDB8 A0A0A1XJI6 A0A0N0BCT9 A0A182MJS5 K7ISQ6 A0A182WJQ7 A0A151XHN3 A0A182XLK9 A0A182PTY0 A0A182R3I0 A0A182JPM6 A0A0L7QX38 A0A1B0G8G3 A0A310SEP7 A0A232F4G8 A0A3B0KFL9 A0A0J7L8E5
A0A194PYB4 A0A0L7LV38 A0A0K0Q0G3 D6WFS4 A0A1Y1KP07 A0A1W4XC42 N6U3L9 V5GJ36 A0A0T6BC77 Q7QJ29 A0A1B6E548 A0A1B6EDT6 A0A1B6DAD0 A0A1B6CFM4 A0A1B6DJE2 A0A1J1IUS8 A0A084VQX4 Q2UZU6 A0A1B6K977 B4KYM2 A0A182UXG5 A0A182QQU6 A0A2C9GPS4 A0A2P8Y7Y4 B4N392 W5JXD0 A0A0M3QW13 D2NUH5 Q9VZZ4 B4H1J9 Q29FB4 E0R979 A0A182KQP7 B3NBK0 B4IZC8 A0A0J9RML2 B4QNK1 A0A3B0KKT0 A0A1W4VP18 A0A1S4G837 E0VZU0 B3M7Y3 A0A2S2QJH4 B4PDD3 A0A2S2NP02 A0A2H8TLB2 A0A0L0C335 B4LDU2 A0A1I8MZ49 T1PGN3 A0A1I8QDU7 A0A0N7ZAC2 A0A0P4VSH7 A0A2P2HZE6 T1DFR8 A0A0P4VX44 A0A0P6IDE8 A0A0P6DAX5 A0A0P5ZY75 A0A0N8EFR5 A0A0P6GUX0 A0A0P5GP60 A0A0P5M4M6 A0A0N7ZKD4 A0A2A3EPZ0 A0A088A0S4 A0A0P6BZV6 Q17PE4 A0A034VZ39 A0A2L2YGD2 A0A2L2YGP6 A0A1A9V3P3 A0A2S2NNG2 A0A0C9QK01 A0A0C9Q881 A0A087U8G0 W8B783 A0A182TDB8 A0A0A1XJI6 A0A0N0BCT9 A0A182MJS5 K7ISQ6 A0A182WJQ7 A0A151XHN3 A0A182XLK9 A0A182PTY0 A0A182R3I0 A0A182JPM6 A0A0L7QX38 A0A1B0G8G3 A0A310SEP7 A0A232F4G8 A0A3B0KFL9 A0A0J7L8E5
PDB
6BMT
E-value=3.69207e-134,
Score=1230
Ontologies
GO
PANTHER
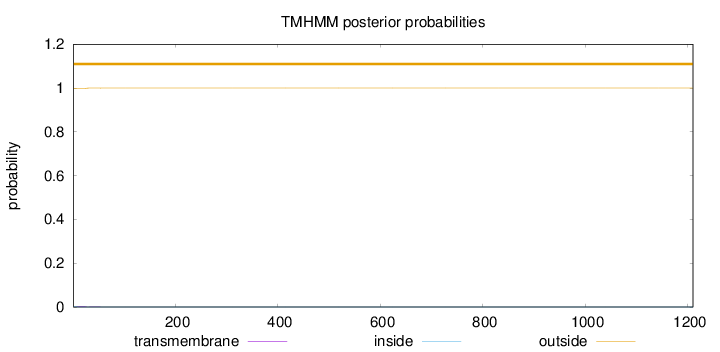
Topology
Subcellular location
Secreted
Length:
1210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0366900000000001
Exp number, first 60 AAs:
0.03535
Total prob of N-in:
0.00195
outside
1 - 1210
Population Genetic Test Statistics
Pi
19.214079
Theta
22.205734
Tajima's D
-0.627676
CLR
0.123329
CSRT
0.215089245537723
Interpretation
Uncertain
