Gene
KWMTBOMO00266
Pre Gene Modal
BGIBMGA000666
Annotation
CRAL-TRIO_domain-containing_protein?_partial_[Manduca_sexta]
Location in the cell
Nuclear Reliability : 2.496
Sequence
CDS
ATGACGACGGCCAGCGTTCTATTAACGGTGACTACTGCCTGGAATGTGACAATCGAGAAAGCAGAGAATGACGGTAAGATGGGACGAACTCTAAAAAATGTGGTCGCCGAAGCCATCACAGAACTTCGAGAAACTCAGTCTACTAAGGAGCAATCTCTTCAACTTATGAGAGAGTGGATAGAACAGAATGGAGACATCAAAAACGTACGACAAGATGAAATATTTTTACTACGATTTCTGCGACATAAGAAGTATAGCATACCAATGGCCCAACAAACTTTACTTAAATATTTAAGTCTCAGAAAGTTCTACCCCGAAATTTTTAAAAATTTGGATTACGAGGATAGCAACTTACAAGAAATTATAAATAATGGGTACATAGTGGTTTCACCAGTTCGTGACAACAAGGGTCGCCGGGTCATCTTTTACAATATGAGTAAATTCAACGCCACTAGATTCAGTTGTTGGGATATGTGCCGTGCTCATATGCTTGTATATGAGAGTTTAATGGATGATCCATTAAATCAAATATGTGGGTTTAATCACATTGGCGATGGAAATGGAATTTCTGGAGCTCACATCACTGCTTGGAACCCCAGCGATTTCGGAAGGATTATAAAATGGGGGGAGCAATCGTTGTCGATGCGACACAAAGCGGTTCATCTGATAAACGTTCCCGTCGCTCTCAAATACGTCATAGACTTCGCAACATCTAAAGTGTCACAGAAAATGGCACATAGACTAAAAGTATGCACAAACTCAAAACTATTACATACACAAACAGACGTTGACTGTCTACCCACTTTATACGGAGGCAAATTAGAATTGCGTGAAATGATAGAATACACCAAGCAAATTTTGAAAGAACAAAAAGAAAATGTTCTTGCTTTGGACAACATGGAGATCCTCAGCACACGTGGGATAATTTCATCGAGAGGCAGGAACCGAGATAACAATGACAATGAAGATTTAATTTCAGTCGAAGGCAGTTTCAGAAAACTTGAAATAGATTAA
Protein
MTTASVLLTVTTAWNVTIEKAENDGKMGRTLKNVVAEAITELRETQSTKEQSLQLMREWIEQNGDIKNVRQDEIFLLRFLRHKKYSIPMAQQTLLKYLSLRKFYPEIFKNLDYEDSNLQEIINNGYIVVSPVRDNKGRRVIFYNMSKFNATRFSCWDMCRAHMLVYESLMDDPLNQICGFNHIGDGNGISGAHITAWNPSDFGRIIKWGEQSLSMRHKAVHLINVPVALKYVIDFATSKVSQKMAHRLKVCTNSKLLHTQTDVDCLPTLYGGKLELREMIEYTKQILKEQKENVLALDNMEILSTRGIISSRGRNRDNNDNEDLISVEGSFRKLEID
Summary
Uniprot
A0A0S2Z3N7
A0A2H1V4Q1
A0A2A4JBY7
A0A194QUU2
A0A2H4RMQ1
A0A212EXX4
+ More
A0A2H4RMV7 A0A194PYB9 A0A0L7LV34 A0A1I8MS85 A0A1L8DZ54 A0A0L0CPL4 A0A182NM49 A0A182QY39 A0A182FCH8 A0A182MSP2 A0A2M3ZAZ2 A0A182U0G7 B0WNM0 A0A182PW99 A0A0A1XGL8 A0A1Q3F051 A0A182WKE3 A0A182VN78 A0A182LJD7 A0A182X534 Q7Q719 A0A182HLY1 A0A182Y124 W5JVA9 A0A182RMG5 B4KXK0 A0A0K8UGY8 B4IXE5 A0A034VEB7 A0A084WKL1 B4HAB9 Q2LZB5 A0A182KBB9 A0A0M4EJY4 B4MLQ7 B4LG43 A0A3B0KJ18 A0A1I8NNZ2 A0A1B0D0M1 A0A0J9RR13 A0A1W4UWZ8 A0A069DTA5 B3M3M1 B4PIX3 B4HUJ0 W8B9M2 A0A023F317 Q16PJ3 A0A182ILW2 B3NC91 Q9VRP8 A0A182G9R3 A0A182H5L3 A0A023EPM6 A0A1B6IYA5 N6U3C1 A0A0K8SQ13 A0A2J7QIH9 A0A1B6HID8 T1GSY0 A0A2S2PB09 A0A0A9WJV2 A0A2P8XVF4 A0A067RPT1 A0A1A9VDV5 A0A2H8TN62 A0A1B0FK05 A0A1B6DJD5 A0A1A9ZWI2 A0A1B0C7D8 J9K2X8 A0A1A9W4K8 A0A139WKP5 A0A336LZV6 A0A0T6B0U3 A0A139WKD4 A0A139WK97 A0A2S2QG74 A0A1J1HZN0 A0A023F2I7 E9I8F2 A0A023F3G8 A0A1B6FJZ8 A0A224XSK7 Q16EW5 E2AKP8 A0A1B6JQG9 A0A026X4A6 E2BVA7 A0A0L7QSH9 A0A0N0U5W8
A0A2H4RMV7 A0A194PYB9 A0A0L7LV34 A0A1I8MS85 A0A1L8DZ54 A0A0L0CPL4 A0A182NM49 A0A182QY39 A0A182FCH8 A0A182MSP2 A0A2M3ZAZ2 A0A182U0G7 B0WNM0 A0A182PW99 A0A0A1XGL8 A0A1Q3F051 A0A182WKE3 A0A182VN78 A0A182LJD7 A0A182X534 Q7Q719 A0A182HLY1 A0A182Y124 W5JVA9 A0A182RMG5 B4KXK0 A0A0K8UGY8 B4IXE5 A0A034VEB7 A0A084WKL1 B4HAB9 Q2LZB5 A0A182KBB9 A0A0M4EJY4 B4MLQ7 B4LG43 A0A3B0KJ18 A0A1I8NNZ2 A0A1B0D0M1 A0A0J9RR13 A0A1W4UWZ8 A0A069DTA5 B3M3M1 B4PIX3 B4HUJ0 W8B9M2 A0A023F317 Q16PJ3 A0A182ILW2 B3NC91 Q9VRP8 A0A182G9R3 A0A182H5L3 A0A023EPM6 A0A1B6IYA5 N6U3C1 A0A0K8SQ13 A0A2J7QIH9 A0A1B6HID8 T1GSY0 A0A2S2PB09 A0A0A9WJV2 A0A2P8XVF4 A0A067RPT1 A0A1A9VDV5 A0A2H8TN62 A0A1B0FK05 A0A1B6DJD5 A0A1A9ZWI2 A0A1B0C7D8 J9K2X8 A0A1A9W4K8 A0A139WKP5 A0A336LZV6 A0A0T6B0U3 A0A139WKD4 A0A139WK97 A0A2S2QG74 A0A1J1HZN0 A0A023F2I7 E9I8F2 A0A023F3G8 A0A1B6FJZ8 A0A224XSK7 Q16EW5 E2AKP8 A0A1B6JQG9 A0A026X4A6 E2BVA7 A0A0L7QSH9 A0A0N0U5W8
Pubmed
25684408
26354079
29136137
22118469
26227816
25315136
+ More
26108605 25830018 20966253 12364791 14747013 17210077 25244985 20920257 23761445 17994087 18057021 25348373 24438588 15632085 22936249 26334808 17550304 24495485 25474469 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 24945155 23537049 26823975 25401762 29403074 24845553 18362917 19820115 21282665 20798317 24508170
26108605 25830018 20966253 12364791 14747013 17210077 25244985 20920257 23761445 17994087 18057021 25348373 24438588 15632085 22936249 26334808 17550304 24495485 25474469 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26483478 24945155 23537049 26823975 25401762 29403074 24845553 18362917 19820115 21282665 20798317 24508170
EMBL
KT943563
ALQ33321.1
ODYU01000655
SOQ35807.1
NWSH01002204
PCG68913.1
+ More
KQ461108 KPJ09237.1 MG434611 ATY51917.1 AGBW02011660 OWR46340.1 MG434661 ATY51967.1 KQ459594 KPI96145.1 JTDY01000029 KOB79312.1 GFDF01002346 JAV11738.1 JRES01000080 KNC34318.1 AXCN02000985 AXCM01000863 GGFM01004921 MBW25672.1 DS232013 EDS31856.1 GBXI01003758 JAD10534.1 GFDL01014118 JAV20927.1 AAAB01008960 EAA11343.4 APCN01004272 ADMH02000008 ETN68116.1 CH933809 EDW18686.1 KRG06316.1 GDHF01026367 GDHF01011649 JAI25947.1 JAI40665.1 CH916366 EDV96382.1 GAKP01018116 GAKP01018115 GAKP01018113 GAKP01018110 JAC40839.1 ATLV01024125 KE525349 KFB50755.1 CH479240 EDW37517.1 CH379069 EAL29593.2 CP012525 ALC43422.1 CH963847 EDW72983.2 CH940647 EDW69351.1 KRF84334.1 OUUW01000009 SPP85081.1 AJVK01009964 AJVK01009965 CM002912 KMY97799.1 GBGD01002017 JAC86872.1 CH902618 EDV40314.1 CM000159 EDW93543.1 CH480817 EDW50611.1 GAMC01020086 JAB86469.1 GBBI01003206 JAC15506.1 CH477779 EAT36298.1 CH954178 EDV51049.1 AE014296 AAF50745.1 JXUM01008310 KQ560257 KXJ83518.1 JXUM01027246 KQ560784 KXJ80897.1 GAPW01002620 JAC10978.1 GECU01015789 JAS91917.1 APGK01041377 KB740993 KB632256 ENN76065.1 ERL90716.1 GBRD01010456 GDHC01007132 JAG55368.1 JAQ11497.1 NEVH01013592 PNF28391.1 GECU01033278 JAS74428.1 CAQQ02114471 GGMR01013955 MBY26574.1 GBHO01036811 JAG06793.1 PYGN01001288 PSN35983.1 KK852542 KDR21724.1 GFXV01003654 MBW15459.1 CCAG010003334 GEDC01011553 JAS25745.1 JXJN01028264 ABLF02033376 KQ971328 KYB28391.1 UFQT01000211 SSX21757.1 LJIG01016276 KRT81087.1 KYB28392.1 KYB28390.1 GGMS01007533 MBY76736.1 CVRI01000035 CRK92780.1 GBBI01002885 JAC15827.1 GL761538 EFZ23179.1 GBBI01002886 JAC15826.1 GECZ01019268 JAS50501.1 GFTR01004914 JAW11512.1 CH478564 EAT32778.1 GL440368 EFN65988.1 GECU01006286 JAT01421.1 KK107015 EZA62861.1 GL450824 EFN80399.1 KQ414758 KOC61504.1 KQ435752 KOX76208.1
KQ461108 KPJ09237.1 MG434611 ATY51917.1 AGBW02011660 OWR46340.1 MG434661 ATY51967.1 KQ459594 KPI96145.1 JTDY01000029 KOB79312.1 GFDF01002346 JAV11738.1 JRES01000080 KNC34318.1 AXCN02000985 AXCM01000863 GGFM01004921 MBW25672.1 DS232013 EDS31856.1 GBXI01003758 JAD10534.1 GFDL01014118 JAV20927.1 AAAB01008960 EAA11343.4 APCN01004272 ADMH02000008 ETN68116.1 CH933809 EDW18686.1 KRG06316.1 GDHF01026367 GDHF01011649 JAI25947.1 JAI40665.1 CH916366 EDV96382.1 GAKP01018116 GAKP01018115 GAKP01018113 GAKP01018110 JAC40839.1 ATLV01024125 KE525349 KFB50755.1 CH479240 EDW37517.1 CH379069 EAL29593.2 CP012525 ALC43422.1 CH963847 EDW72983.2 CH940647 EDW69351.1 KRF84334.1 OUUW01000009 SPP85081.1 AJVK01009964 AJVK01009965 CM002912 KMY97799.1 GBGD01002017 JAC86872.1 CH902618 EDV40314.1 CM000159 EDW93543.1 CH480817 EDW50611.1 GAMC01020086 JAB86469.1 GBBI01003206 JAC15506.1 CH477779 EAT36298.1 CH954178 EDV51049.1 AE014296 AAF50745.1 JXUM01008310 KQ560257 KXJ83518.1 JXUM01027246 KQ560784 KXJ80897.1 GAPW01002620 JAC10978.1 GECU01015789 JAS91917.1 APGK01041377 KB740993 KB632256 ENN76065.1 ERL90716.1 GBRD01010456 GDHC01007132 JAG55368.1 JAQ11497.1 NEVH01013592 PNF28391.1 GECU01033278 JAS74428.1 CAQQ02114471 GGMR01013955 MBY26574.1 GBHO01036811 JAG06793.1 PYGN01001288 PSN35983.1 KK852542 KDR21724.1 GFXV01003654 MBW15459.1 CCAG010003334 GEDC01011553 JAS25745.1 JXJN01028264 ABLF02033376 KQ971328 KYB28391.1 UFQT01000211 SSX21757.1 LJIG01016276 KRT81087.1 KYB28392.1 KYB28390.1 GGMS01007533 MBY76736.1 CVRI01000035 CRK92780.1 GBBI01002885 JAC15827.1 GL761538 EFZ23179.1 GBBI01002886 JAC15826.1 GECZ01019268 JAS50501.1 GFTR01004914 JAW11512.1 CH478564 EAT32778.1 GL440368 EFN65988.1 GECU01006286 JAT01421.1 KK107015 EZA62861.1 GL450824 EFN80399.1 KQ414758 KOC61504.1 KQ435752 KOX76208.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
UP000095301
+ More
UP000037069 UP000075884 UP000075886 UP000069272 UP000075883 UP000075902 UP000002320 UP000075885 UP000075920 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000000673 UP000075900 UP000009192 UP000001070 UP000030765 UP000008744 UP000001819 UP000075881 UP000092553 UP000007798 UP000008792 UP000268350 UP000095300 UP000092462 UP000192221 UP000007801 UP000002282 UP000001292 UP000008820 UP000075880 UP000008711 UP000000803 UP000069940 UP000249989 UP000019118 UP000030742 UP000235965 UP000015102 UP000245037 UP000027135 UP000078200 UP000092444 UP000092445 UP000092460 UP000007819 UP000091820 UP000007266 UP000183832 UP000000311 UP000053097 UP000008237 UP000053825 UP000053105
UP000037069 UP000075884 UP000075886 UP000069272 UP000075883 UP000075902 UP000002320 UP000075885 UP000075920 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000076408 UP000000673 UP000075900 UP000009192 UP000001070 UP000030765 UP000008744 UP000001819 UP000075881 UP000092553 UP000007798 UP000008792 UP000268350 UP000095300 UP000092462 UP000192221 UP000007801 UP000002282 UP000001292 UP000008820 UP000075880 UP000008711 UP000000803 UP000069940 UP000249989 UP000019118 UP000030742 UP000235965 UP000015102 UP000245037 UP000027135 UP000078200 UP000092444 UP000092445 UP000092460 UP000007819 UP000091820 UP000007266 UP000183832 UP000000311 UP000053097 UP000008237 UP000053825 UP000053105
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0S2Z3N7
A0A2H1V4Q1
A0A2A4JBY7
A0A194QUU2
A0A2H4RMQ1
A0A212EXX4
+ More
A0A2H4RMV7 A0A194PYB9 A0A0L7LV34 A0A1I8MS85 A0A1L8DZ54 A0A0L0CPL4 A0A182NM49 A0A182QY39 A0A182FCH8 A0A182MSP2 A0A2M3ZAZ2 A0A182U0G7 B0WNM0 A0A182PW99 A0A0A1XGL8 A0A1Q3F051 A0A182WKE3 A0A182VN78 A0A182LJD7 A0A182X534 Q7Q719 A0A182HLY1 A0A182Y124 W5JVA9 A0A182RMG5 B4KXK0 A0A0K8UGY8 B4IXE5 A0A034VEB7 A0A084WKL1 B4HAB9 Q2LZB5 A0A182KBB9 A0A0M4EJY4 B4MLQ7 B4LG43 A0A3B0KJ18 A0A1I8NNZ2 A0A1B0D0M1 A0A0J9RR13 A0A1W4UWZ8 A0A069DTA5 B3M3M1 B4PIX3 B4HUJ0 W8B9M2 A0A023F317 Q16PJ3 A0A182ILW2 B3NC91 Q9VRP8 A0A182G9R3 A0A182H5L3 A0A023EPM6 A0A1B6IYA5 N6U3C1 A0A0K8SQ13 A0A2J7QIH9 A0A1B6HID8 T1GSY0 A0A2S2PB09 A0A0A9WJV2 A0A2P8XVF4 A0A067RPT1 A0A1A9VDV5 A0A2H8TN62 A0A1B0FK05 A0A1B6DJD5 A0A1A9ZWI2 A0A1B0C7D8 J9K2X8 A0A1A9W4K8 A0A139WKP5 A0A336LZV6 A0A0T6B0U3 A0A139WKD4 A0A139WK97 A0A2S2QG74 A0A1J1HZN0 A0A023F2I7 E9I8F2 A0A023F3G8 A0A1B6FJZ8 A0A224XSK7 Q16EW5 E2AKP8 A0A1B6JQG9 A0A026X4A6 E2BVA7 A0A0L7QSH9 A0A0N0U5W8
A0A2H4RMV7 A0A194PYB9 A0A0L7LV34 A0A1I8MS85 A0A1L8DZ54 A0A0L0CPL4 A0A182NM49 A0A182QY39 A0A182FCH8 A0A182MSP2 A0A2M3ZAZ2 A0A182U0G7 B0WNM0 A0A182PW99 A0A0A1XGL8 A0A1Q3F051 A0A182WKE3 A0A182VN78 A0A182LJD7 A0A182X534 Q7Q719 A0A182HLY1 A0A182Y124 W5JVA9 A0A182RMG5 B4KXK0 A0A0K8UGY8 B4IXE5 A0A034VEB7 A0A084WKL1 B4HAB9 Q2LZB5 A0A182KBB9 A0A0M4EJY4 B4MLQ7 B4LG43 A0A3B0KJ18 A0A1I8NNZ2 A0A1B0D0M1 A0A0J9RR13 A0A1W4UWZ8 A0A069DTA5 B3M3M1 B4PIX3 B4HUJ0 W8B9M2 A0A023F317 Q16PJ3 A0A182ILW2 B3NC91 Q9VRP8 A0A182G9R3 A0A182H5L3 A0A023EPM6 A0A1B6IYA5 N6U3C1 A0A0K8SQ13 A0A2J7QIH9 A0A1B6HID8 T1GSY0 A0A2S2PB09 A0A0A9WJV2 A0A2P8XVF4 A0A067RPT1 A0A1A9VDV5 A0A2H8TN62 A0A1B0FK05 A0A1B6DJD5 A0A1A9ZWI2 A0A1B0C7D8 J9K2X8 A0A1A9W4K8 A0A139WKP5 A0A336LZV6 A0A0T6B0U3 A0A139WKD4 A0A139WK97 A0A2S2QG74 A0A1J1HZN0 A0A023F2I7 E9I8F2 A0A023F3G8 A0A1B6FJZ8 A0A224XSK7 Q16EW5 E2AKP8 A0A1B6JQG9 A0A026X4A6 E2BVA7 A0A0L7QSH9 A0A0N0U5W8
PDB
3HY5
E-value=6.41136e-18,
Score=222
Ontologies
GO
Topology
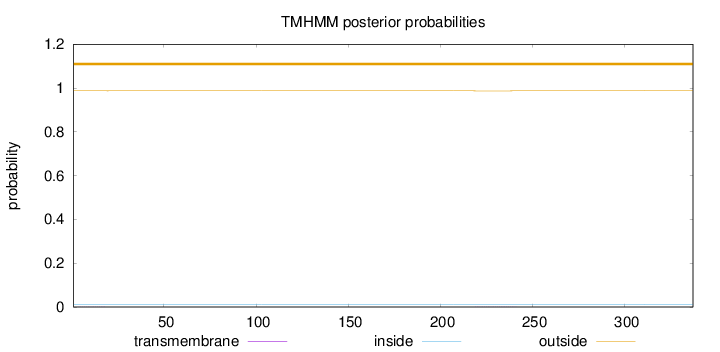
Length:
337
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01595
Exp number, first 60 AAs:
0.00235
Total prob of N-in:
0.01274
outside
1 - 337
Population Genetic Test Statistics
Pi
15.154894
Theta
18.81422
Tajima's D
-0.760799
CLR
0.036059
CSRT
0.181840907954602
Interpretation
Uncertain
