Gene
KWMTBOMO00261
Annotation
PREDICTED:_uncharacterized_protein_LOC106133776_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.279
Sequence
CDS
ATGGTGCCCACTATTTACTATGAATTTTCGCAAGCCCAGTTGAGACTTGGATCTTATGAAAGCTGTGACAAAACATTCTTCCGGCATTATAGAGATAAAATTCACGAACATTGTCTAGTGGCCGTTAAAACGCATTGCCACAACATATCTAATTTAAAGGTCATATTTGCTATAATTTGCTCAATAGTATTGGAAGTACCTTGTGGATTGACAGCCGCAATGGCAGCCTGTTTATGTATGGAAATCCAAGATTATGCCCTCAACGAAGAAAATTTAGTTGCGTCAAGCCGTTATTGGATGCACGCAATAGTGATTTCAGTGATGTCGCTTATCTGCTGGGTTCATAAAGCTTCGGTGCTTTATCGTTACGTAAATCAAGTTATTTCAAGAAGAGCAAAAGAGGCACCGCATCTCAATCCGCCTCTAATGCAATCTTACAAAATTGGCCATGGTCATGTCACTTGGAATAAACCTACACTTTTTTTTGAAGATTGGGAAATGCGTTTTGGCTTATGGAAGCACTTTAAAGATGCCCAGCCAATTACAGGAAATAAAGCTTAA
Protein
MVPTIYYEFSQAQLRLGSYESCDKTFFRHYRDKIHEHCLVAVKTHCHNISNLKVIFAIICSIVLEVPCGLTAAMAACLCMEIQDYALNEENLVASSRYWMHAIVISVMSLICWVHKASVLYRYVNQVISRRAKEAPHLNPPLMQSYKIGHGHVTWNKPTLFFEDWEMRFGLWKHFKDAQPITGNKA
Summary
Uniprot
A0A2A4JBW2
A0A2W1BMN4
A0A2H1V4N8
A0A212EXJ4
A0A0L7QNH7
A0A310S7D2
+ More
F4WPV6 A0A087ZZI3 A0A0J7L8R8 E2AC07 A0A151WR46 A0A154PIC1 A0A195AX47 A0A195FEE3 D6WVV5 A0A195EH45 A0A139WDL1 A0A0M9A4Z0 E9IBL5 A0A195CKS9 A0A3L8D8K3 A0A026WZ15 A0A158NI95 E2BXG6 A0A232F7D0 K7JRN3 U4U5Y4 A0A182GIE8 N6UB44 A0A182NFW8 A0A084VDK2 F5HJB7 A0A182QA38 A0A182TXJ0 A0A182LAN8 A0A182I8S4 A0A182UZB7 A0A182WV50 A0A182SRX4 A0A182R5X9 A0A182KDU8 A0A182ILW5 A0A182VXU4 A0A182XXD1 A0A182P778 A0A182MH57 A0A182F9G0 A0A1J1J2N4 D6WPQ4 A0A0M9A5I6 A0A0T6B617 A0A0L7QVC9 A0A310SQ45 A0A1Y1MSW0 A0A088A2T2 A0A182MXA5 K7J5Z4 A0A2A3ECQ4 A0A212EU68 A0A182QAK5 A0A232ETW7 A0A182K9K0 A0A182RAK3 A0A182MHC7 A0A182PGD5 A0A067R5J2 A0A147ATC0 A0A146Y9R2 A0A2M3Z6G8 A0A182KSZ5 A0A182HPT9 Q5TN25 A0A182UXC0 A0A1D6T371 A0A182TL03 A0A182WZZ0 A0A3P8SDX3 A0A182FVM8 A0A182JL92
F4WPV6 A0A087ZZI3 A0A0J7L8R8 E2AC07 A0A151WR46 A0A154PIC1 A0A195AX47 A0A195FEE3 D6WVV5 A0A195EH45 A0A139WDL1 A0A0M9A4Z0 E9IBL5 A0A195CKS9 A0A3L8D8K3 A0A026WZ15 A0A158NI95 E2BXG6 A0A232F7D0 K7JRN3 U4U5Y4 A0A182GIE8 N6UB44 A0A182NFW8 A0A084VDK2 F5HJB7 A0A182QA38 A0A182TXJ0 A0A182LAN8 A0A182I8S4 A0A182UZB7 A0A182WV50 A0A182SRX4 A0A182R5X9 A0A182KDU8 A0A182ILW5 A0A182VXU4 A0A182XXD1 A0A182P778 A0A182MH57 A0A182F9G0 A0A1J1J2N4 D6WPQ4 A0A0M9A5I6 A0A0T6B617 A0A0L7QVC9 A0A310SQ45 A0A1Y1MSW0 A0A088A2T2 A0A182MXA5 K7J5Z4 A0A2A3ECQ4 A0A212EU68 A0A182QAK5 A0A232ETW7 A0A182K9K0 A0A182RAK3 A0A182MHC7 A0A182PGD5 A0A067R5J2 A0A147ATC0 A0A146Y9R2 A0A2M3Z6G8 A0A182KSZ5 A0A182HPT9 Q5TN25 A0A182UXC0 A0A1D6T371 A0A182TL03 A0A182WZZ0 A0A3P8SDX3 A0A182FVM8 A0A182JL92
Pubmed
EMBL
NWSH01002204
PCG68920.1
KZ149961
PZC76319.1
ODYU01000655
SOQ35801.1
+ More
AGBW02011730 OWR46213.1 KQ414855 KOC60124.1 KQ779356 OAD51999.1 GL888255 EGI63785.1 LBMM01000249 KMR03011.1 GL438389 EFN69025.1 KQ982813 KYQ50336.1 KQ434924 KZC11583.1 KQ976725 KYM76534.1 KQ981636 KYN38773.1 KQ971358 EFA08236.2 KQ978957 KYN27197.1 KYB25992.1 KQ435732 KOX77335.1 GL762111 EFZ22135.1 KQ977622 KYN01313.1 QOIP01000011 RLU16825.1 KK107078 EZA60384.1 ADTU01016114 GL451254 EFN79614.1 NNAY01000815 OXU26359.1 KB632064 ERL88477.1 JXUM01065891 KQ562371 KXJ76049.1 APGK01035623 KB740928 KI210309 ENN77876.1 ERL96238.1 ATLV01011572 KE524696 KFB36046.1 AAAB01008844 EGK96378.1 AXCN02001148 APCN01003108 AXCM01006855 CVRI01000067 CRL06632.1 KQ971354 EFA06189.2 KQ435750 KOX76289.1 LJIG01009607 KRT82742.1 KQ414731 KOC62431.1 KQ760240 OAD61313.1 GEZM01023738 JAV88208.1 KZ288293 PBC28989.1 AGBW02012452 OWR45040.1 AXCN02001678 NNAY01002205 OXU21810.1 AXCM01008745 KK852686 KDR18451.1 GCES01036057 GCES01004652 JAR81671.1 GCES01036061 JAR50262.1 GGFM01003372 MBW24123.1 APCN01003271 AAAB01008986 EAL38737.3
AGBW02011730 OWR46213.1 KQ414855 KOC60124.1 KQ779356 OAD51999.1 GL888255 EGI63785.1 LBMM01000249 KMR03011.1 GL438389 EFN69025.1 KQ982813 KYQ50336.1 KQ434924 KZC11583.1 KQ976725 KYM76534.1 KQ981636 KYN38773.1 KQ971358 EFA08236.2 KQ978957 KYN27197.1 KYB25992.1 KQ435732 KOX77335.1 GL762111 EFZ22135.1 KQ977622 KYN01313.1 QOIP01000011 RLU16825.1 KK107078 EZA60384.1 ADTU01016114 GL451254 EFN79614.1 NNAY01000815 OXU26359.1 KB632064 ERL88477.1 JXUM01065891 KQ562371 KXJ76049.1 APGK01035623 KB740928 KI210309 ENN77876.1 ERL96238.1 ATLV01011572 KE524696 KFB36046.1 AAAB01008844 EGK96378.1 AXCN02001148 APCN01003108 AXCM01006855 CVRI01000067 CRL06632.1 KQ971354 EFA06189.2 KQ435750 KOX76289.1 LJIG01009607 KRT82742.1 KQ414731 KOC62431.1 KQ760240 OAD61313.1 GEZM01023738 JAV88208.1 KZ288293 PBC28989.1 AGBW02012452 OWR45040.1 AXCN02001678 NNAY01002205 OXU21810.1 AXCM01008745 KK852686 KDR18451.1 GCES01036057 GCES01004652 JAR81671.1 GCES01036061 JAR50262.1 GGFM01003372 MBW24123.1 APCN01003271 AAAB01008986 EAL38737.3
Proteomes
UP000218220
UP000007151
UP000053825
UP000007755
UP000005203
UP000036403
+ More
UP000000311 UP000075809 UP000076502 UP000078540 UP000078541 UP000007266 UP000078492 UP000053105 UP000078542 UP000279307 UP000053097 UP000005205 UP000008237 UP000215335 UP000002358 UP000030742 UP000069940 UP000249989 UP000019118 UP000075884 UP000030765 UP000007062 UP000075886 UP000075902 UP000075882 UP000075840 UP000075903 UP000076407 UP000075901 UP000075900 UP000075881 UP000075880 UP000075920 UP000076408 UP000075885 UP000075883 UP000069272 UP000183832 UP000242457 UP000027135 UP000265080
UP000000311 UP000075809 UP000076502 UP000078540 UP000078541 UP000007266 UP000078492 UP000053105 UP000078542 UP000279307 UP000053097 UP000005205 UP000008237 UP000215335 UP000002358 UP000030742 UP000069940 UP000249989 UP000019118 UP000075884 UP000030765 UP000007062 UP000075886 UP000075902 UP000075882 UP000075840 UP000075903 UP000076407 UP000075901 UP000075900 UP000075881 UP000075880 UP000075920 UP000076408 UP000075885 UP000075883 UP000069272 UP000183832 UP000242457 UP000027135 UP000265080
Interpro
IPR022591
TFIID_sub1_DUF3591
+ More
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
IPR040240 TAF1
IPR036427 Bromodomain-like_sf
IPR000210 BTB/POZ_dom
IPR036236 Znf_C2H2_sf
IPR011333 SKP1/BTB/POZ_sf
IPR036741 TAFII-230_TBP-bd_sf
IPR013087 Znf_C2H2_type
IPR041670 Znf-CCHC_6
IPR009067 TAF_II_230-bd
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR001313 Pumilio_RNA-bd_rpt
IPR018359 Bromodomain_CS
IPR001487 Bromodomain
IPR040240 TAF1
IPR036427 Bromodomain-like_sf
IPR000210 BTB/POZ_dom
IPR036236 Znf_C2H2_sf
IPR011333 SKP1/BTB/POZ_sf
IPR036741 TAFII-230_TBP-bd_sf
IPR013087 Znf_C2H2_type
IPR041670 Znf-CCHC_6
IPR009067 TAF_II_230-bd
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR001313 Pumilio_RNA-bd_rpt
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JBW2
A0A2W1BMN4
A0A2H1V4N8
A0A212EXJ4
A0A0L7QNH7
A0A310S7D2
+ More
F4WPV6 A0A087ZZI3 A0A0J7L8R8 E2AC07 A0A151WR46 A0A154PIC1 A0A195AX47 A0A195FEE3 D6WVV5 A0A195EH45 A0A139WDL1 A0A0M9A4Z0 E9IBL5 A0A195CKS9 A0A3L8D8K3 A0A026WZ15 A0A158NI95 E2BXG6 A0A232F7D0 K7JRN3 U4U5Y4 A0A182GIE8 N6UB44 A0A182NFW8 A0A084VDK2 F5HJB7 A0A182QA38 A0A182TXJ0 A0A182LAN8 A0A182I8S4 A0A182UZB7 A0A182WV50 A0A182SRX4 A0A182R5X9 A0A182KDU8 A0A182ILW5 A0A182VXU4 A0A182XXD1 A0A182P778 A0A182MH57 A0A182F9G0 A0A1J1J2N4 D6WPQ4 A0A0M9A5I6 A0A0T6B617 A0A0L7QVC9 A0A310SQ45 A0A1Y1MSW0 A0A088A2T2 A0A182MXA5 K7J5Z4 A0A2A3ECQ4 A0A212EU68 A0A182QAK5 A0A232ETW7 A0A182K9K0 A0A182RAK3 A0A182MHC7 A0A182PGD5 A0A067R5J2 A0A147ATC0 A0A146Y9R2 A0A2M3Z6G8 A0A182KSZ5 A0A182HPT9 Q5TN25 A0A182UXC0 A0A1D6T371 A0A182TL03 A0A182WZZ0 A0A3P8SDX3 A0A182FVM8 A0A182JL92
F4WPV6 A0A087ZZI3 A0A0J7L8R8 E2AC07 A0A151WR46 A0A154PIC1 A0A195AX47 A0A195FEE3 D6WVV5 A0A195EH45 A0A139WDL1 A0A0M9A4Z0 E9IBL5 A0A195CKS9 A0A3L8D8K3 A0A026WZ15 A0A158NI95 E2BXG6 A0A232F7D0 K7JRN3 U4U5Y4 A0A182GIE8 N6UB44 A0A182NFW8 A0A084VDK2 F5HJB7 A0A182QA38 A0A182TXJ0 A0A182LAN8 A0A182I8S4 A0A182UZB7 A0A182WV50 A0A182SRX4 A0A182R5X9 A0A182KDU8 A0A182ILW5 A0A182VXU4 A0A182XXD1 A0A182P778 A0A182MH57 A0A182F9G0 A0A1J1J2N4 D6WPQ4 A0A0M9A5I6 A0A0T6B617 A0A0L7QVC9 A0A310SQ45 A0A1Y1MSW0 A0A088A2T2 A0A182MXA5 K7J5Z4 A0A2A3ECQ4 A0A212EU68 A0A182QAK5 A0A232ETW7 A0A182K9K0 A0A182RAK3 A0A182MHC7 A0A182PGD5 A0A067R5J2 A0A147ATC0 A0A146Y9R2 A0A2M3Z6G8 A0A182KSZ5 A0A182HPT9 Q5TN25 A0A182UXC0 A0A1D6T371 A0A182TL03 A0A182WZZ0 A0A3P8SDX3 A0A182FVM8 A0A182JL92
Ontologies
PANTHER
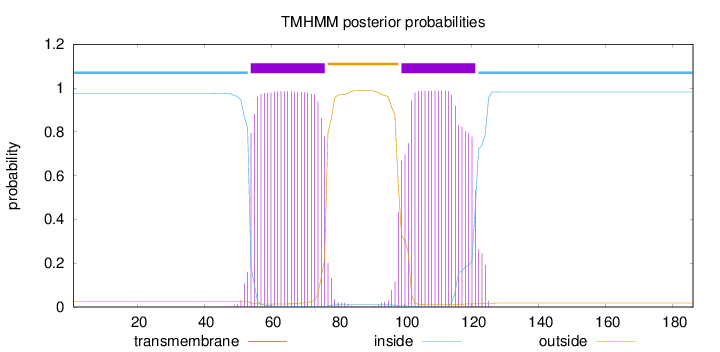
Topology
Length:
186
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.45391
Exp number, first 60 AAs:
6.87196
Total prob of N-in:
0.97594
inside
1 - 53
TMhelix
54 - 76
outside
77 - 98
TMhelix
99 - 121
inside
122 - 186
Population Genetic Test Statistics
Pi
16.004952
Theta
13.438365
Tajima's D
0.61143
CLR
0.533692
CSRT
0.557322133893305
Interpretation
Uncertain
