Gene
KWMTBOMO00256 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000661
Annotation
PREDICTED:_protein_disulfide-isomerase_A5_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.227 ER Reliability : 1.758
Sequence
CDS
ATGGTCAGTAAAATGCATTGGACGGTGCTCACATTGATGACCATATCACTAGTGTTATCTTCAGTTGAAACTAAGAAGCAGAAGACTGCAGTTTTATCAGATATAAGTGACATAAAAGATTTTAAAAAGATACTTAGAACTAAAACCAATGTTTTAGTTTTATTTGTCAATGAAATGAAAAAGGCACATAATATGGTAGAGGTATTTAAAGAAACTGCTGATACAATGAGAGGGCAAGCTACGTTAATTTTGATTGATTGTAACAACAGTGACAACAAAAAACTATGTAAGAAGTTGAAAGTATCACCAGAAAAGCCATTCATTTTAAAACACTATAAAGACGGAGACTTCAATAAGGATTATGACAGAAGTGAATCTGTCAGTTCAATGTGCAATTTCCTGCGAGATCCTACTGGAGACTTGCCTTGGGAAGAGGATCCTACAGCTGTTGATATTTATCACCTTCTTGATGGAGAGGCCATTGTGAAATTCTTGAAGAAAGGTCCTGGTTCACACAAGAGATCTATGATAATGTTTTATGCTCCTTGGTGTGGGTACTGTAAATCAATGAAACCTGATTATGTTGAGGCAGCTGGGGATCTCAAGGGCGAGTCAATTTTAGCTGCAATTGATGTAGCTAAGCCTGGAAATTCTAAAATAAGACAGTTGTATAATATAACTGGGTTTCCTACTCTGTTGTATTTTGAGAGGGGCCAGTTTCGCTTTCCATATAATGGTGAAAACAAAAGGCAAGCCATAGTCGAGTTTATGAGAGATCCCGTAGCTCAGCCGAAACAGAAAAAGAAAGACGTTGTTGATGAAAGCTGGGCTCGTGACACTGATGTGGTGCATTTAAATGGGGACAGTTTTGACGCTATTCTAGCAAAGGCGGAACACGCACTGGTCGTATTCTACGCGCCCTGGTGTGGACACTGCAAACGTATTAAACCAGAGTTCGAAAAAGCGGCCACAAAGATAAAGACTGAAAAGATTAACGGAATATTAGCAGCTGTCGATGCTACAAAGGAGCCAGATTTAGCGTCGAGATTTGGTGTCAAGGGATATCCGACTCTGAAATATTTTAATAAAGGCGAATTTAAGTTCGACGCTGGACATGCCCGACAAGAAGACCAGATTATTAGTTTTATCAAGGACCCGCAAGAGCCACCCCCACCCCCACCACCAGAAAAGCCATGGTCCGAGGAAGAGTCTCCCGTGCGCCACCTCGACTCCTCGACTTTCAGAAACACACTCCGGAAAATCAAACACAGCATTGTCATGTTCTACGCGCCATGGTGCGGCCACTGCAAAAGCACCAAGCCTGAATTCGTTAAGGCGGCGGAACAGTTTGCCGATGAACTGATGGTTGCTTTTGGAGCTGTCGATTGTACTCTACATCAGGATTTGTGCGCTAACTACAACGTCAAGGGCTACCCGACCATTAAGTATTTTAGTTACTTTGATAAAGCGGTTAAGGACTACACGGGAGGCCAGAAGGAAGGCGACTTTGTTTCGTTTATACACAATCAAATGGGGACTAAGCAGACTTCACAAAAGGCCCCCAAGACAACCCAGGACGCGGGTTTCGGTGCTAACGTCTTATTGGCCTTCGACGATGACTTTGAGAAGATAATATCTGTGCCCACGCCCACGATTGTCATGTTCTATGCTACATGGTGCAGCCACTGCGTGACAGCGAAGCCGGCCTTCAGCAGACTCGCTACAGCGCTTAAGACGGAAAATGTACCGGTGAAAGCGGTCGCTTTAGAAGCGGCTGAAAATCCGAAAGCGGCTGACTTCGCAGGCATACAAACTCTGCCCACGTTCAAACTGTTTGCGGCCGGAAAATACGTGACCGACTACGACGGAGATCGGTCCACAGAAGACTTGCTCAAATTCTGCAAGAGGCACGCCGAGGTTAAAGACGAACTGTGA
Protein
MVSKMHWTVLTLMTISLVLSSVETKKQKTAVLSDISDIKDFKKILRTKTNVLVLFVNEMKKAHNMVEVFKETADTMRGQATLILIDCNNSDNKKLCKKLKVSPEKPFILKHYKDGDFNKDYDRSESVSSMCNFLRDPTGDLPWEEDPTAVDIYHLLDGEAIVKFLKKGPGSHKRSMIMFYAPWCGYCKSMKPDYVEAAGDLKGESILAAIDVAKPGNSKIRQLYNITGFPTLLYFERGQFRFPYNGENKRQAIVEFMRDPVAQPKQKKKDVVDESWARDTDVVHLNGDSFDAILAKAEHALVVFYAPWCGHCKRIKPEFEKAATKIKTEKINGILAAVDATKEPDLASRFGVKGYPTLKYFNKGEFKFDAGHARQEDQIISFIKDPQEPPPPPPPEKPWSEEESPVRHLDSSTFRNTLRKIKHSIVMFYAPWCGHCKSTKPEFVKAAEQFADELMVAFGAVDCTLHQDLCANYNVKGYPTIKYFSYFDKAVKDYTGGQKEGDFVSFIHNQMGTKQTSQKAPKTTQDAGFGANVLLAFDDDFEKIISVPTPTIVMFYATWCSHCVTAKPAFSRLATALKTENVPVKAVALEAAENPKAADFAGIQTLPTFKLFAAGKYVTDYDGDRSTEDLLKFCKRHAEVKDEL
Summary
Similarity
Belongs to the protein disulfide isomerase family.
Uniprot
H9ITT1
A0A2W1BSY7
A0A2A4JWB4
A0A2A4JVP2
A0A194PT73
A0A212FLP9
+ More
A0A194QUL3 A0A2H1VBI6 A0A0L7L0H7 A0A1Y1K7N1 A0A0T6AYD4 A0A2M4APL7 A0A2M4APM1 A0A2J7QUC2 A0A182VDE1 A0A182FMU9 Q16Z37 A0A182IMN5 A0A023EXG3 D6X529 W5JI60 A0A2M4BH54 A0A2M3ZE94 A0A2M4BH18 A0A182YHC3 A0A182P8D0 A0A182KJS6 A0A182I0Q8 A0A182H400 Q7PM55 A0A182VSY5 A0A182QMP7 A0A067RRK1 A0A1Q3FDK9 A0A1Q3FDU5 A0A1Q3FE39 A0A182TDT2 A0A2P8ZPK7 B0XJ65 A0A1Q3FEB5 A0A1S4H450 A0A1Q3FEJ4 A0A182XB64 A0A182R6V8 A0A336KFI3 U4UPW9 N6U8M7 N6TB44 A0A1Q3FDT6 L7M7V9 A0A1Q3FDD0 A0A2L2Y199 A0A1B0D1K1 A0A2L2Y1B4 A0A182LXR6 L7MCN0 A0A2T7PR44 A0A1Q3FE34 A0A0P5DZK6 A0A0P5NVM7 A0A0P5A1Z0 A0A293MSL5 A0A1B0CY75 A0A0P5Z6H7 A0A084WD29 A0A1Q3FE26 A0A224XK72 A0A0P6CPQ6 A0A0P5B8M0 A0A1E1X7E8 A0A0P5UUR0 A0A0V0G3Q8 A0A0P5X0Y9 A0A182N8Q5 A0A069DW52 A0A0N8C9B4 A0A1Z5KXN5 A0A146M2Y5 A0A0A9YNZ9 T1I3Q3 A0A087UJU7 A0A0K8SZU8 A0A1D2MEP7 A0A0P6I972 A0A0P5XZ73 A0A0P6BYC2 B4P359 A0A0A1WZD6 A0A0P4WKC0 Q960C0 R7UHJ2 Q9VJZ1 B4Q4S8 B3NAX5 B4IEK7 A0A034WLQ1 V5HQ91 V5HL97 A0A0K8VGQ5 A0A0K8U9W2
A0A194QUL3 A0A2H1VBI6 A0A0L7L0H7 A0A1Y1K7N1 A0A0T6AYD4 A0A2M4APL7 A0A2M4APM1 A0A2J7QUC2 A0A182VDE1 A0A182FMU9 Q16Z37 A0A182IMN5 A0A023EXG3 D6X529 W5JI60 A0A2M4BH54 A0A2M3ZE94 A0A2M4BH18 A0A182YHC3 A0A182P8D0 A0A182KJS6 A0A182I0Q8 A0A182H400 Q7PM55 A0A182VSY5 A0A182QMP7 A0A067RRK1 A0A1Q3FDK9 A0A1Q3FDU5 A0A1Q3FE39 A0A182TDT2 A0A2P8ZPK7 B0XJ65 A0A1Q3FEB5 A0A1S4H450 A0A1Q3FEJ4 A0A182XB64 A0A182R6V8 A0A336KFI3 U4UPW9 N6U8M7 N6TB44 A0A1Q3FDT6 L7M7V9 A0A1Q3FDD0 A0A2L2Y199 A0A1B0D1K1 A0A2L2Y1B4 A0A182LXR6 L7MCN0 A0A2T7PR44 A0A1Q3FE34 A0A0P5DZK6 A0A0P5NVM7 A0A0P5A1Z0 A0A293MSL5 A0A1B0CY75 A0A0P5Z6H7 A0A084WD29 A0A1Q3FE26 A0A224XK72 A0A0P6CPQ6 A0A0P5B8M0 A0A1E1X7E8 A0A0P5UUR0 A0A0V0G3Q8 A0A0P5X0Y9 A0A182N8Q5 A0A069DW52 A0A0N8C9B4 A0A1Z5KXN5 A0A146M2Y5 A0A0A9YNZ9 T1I3Q3 A0A087UJU7 A0A0K8SZU8 A0A1D2MEP7 A0A0P6I972 A0A0P5XZ73 A0A0P6BYC2 B4P359 A0A0A1WZD6 A0A0P4WKC0 Q960C0 R7UHJ2 Q9VJZ1 B4Q4S8 B3NAX5 B4IEK7 A0A034WLQ1 V5HQ91 V5HL97 A0A0K8VGQ5 A0A0K8U9W2
Pubmed
19121390
28756777
26354079
22118469
26227816
28004739
+ More
17510324 24945155 18362917 19820115 20920257 23761445 25244985 20966253 26483478 12364791 24845553 29403074 23537049 25576852 26561354 24438588 28503490 26334808 28528879 26823975 25401762 27289101 17994087 17550304 25830018 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25765539
17510324 24945155 18362917 19820115 20920257 23761445 25244985 20966253 26483478 12364791 24845553 29403074 23537049 25576852 26561354 24438588 28503490 26334808 28528879 26823975 25401762 27289101 17994087 17550304 25830018 23254933 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25348373 25765539
EMBL
BABH01006945
KZ149961
PZC76327.1
NWSH01000478
PCG76109.1
PCG76107.1
+ More
KQ459594 KPI96168.1 AGBW02007730 OWR54657.1 KQ461108 KPJ09218.1 ODYU01001651 SOQ38171.1 JTDY01003791 KOB69003.1 GEZM01095270 JAV55436.1 LJIG01022528 KRT80104.1 GGFK01009398 MBW42719.1 GGFK01009399 MBW42720.1 NEVH01011186 PNF32184.1 CH477499 EAT39913.1 GAPW01000479 JAC13119.1 KQ971382 EEZ97186.1 ADMH02001180 ETN63816.1 GGFJ01003213 MBW52354.1 GGFM01006050 MBW26801.1 GGFJ01003209 MBW52350.1 APCN01005234 JXUM01025946 KQ560739 KXJ81065.1 AAAB01008980 EAA14324.4 AXCN02001858 KK852470 KDR23255.1 GFDL01009390 JAV25655.1 GFDL01009314 JAV25731.1 GFDL01009267 JAV25778.1 PYGN01000002 PSN58434.1 DS233443 EDS30028.1 GFDL01009152 JAV25893.1 GFDL01009097 JAV25948.1 UFQS01000396 UFQS01001481 UFQT01000396 UFQT01001481 SSX03604.1 SSX11169.1 SSX23969.1 KB632309 ERL92070.1 APGK01044530 APGK01044531 KB741026 ENN74957.1 ENN74958.1 GFDL01009387 JAV25658.1 GACK01005077 JAA59957.1 GFDL01009469 JAV25576.1 IAAA01005327 LAA01943.1 AJVK01010273 AJVK01010274 AJVK01010275 AJVK01010276 IAAA01005328 LAA01944.1 AXCM01000229 GACK01003482 JAA61552.1 PZQS01000002 PVD35860.1 GFDL01009231 JAV25814.1 GDIP01150040 JAJ73362.1 GDIQ01137071 JAL14655.1 GDIP01217568 JAJ05834.1 GFWV01019155 MAA43883.1 AJWK01035530 GDIP01166679 GDIP01050394 LRGB01000024 JAM53321.1 KZS21530.1 ATLV01022925 KE525338 KFB48123.1 GFDL01009248 JAV25797.1 GFTR01007576 JAW08850.1 GDIQ01089501 JAN05236.1 GDIP01187928 JAJ35474.1 GFAC01003991 JAT95197.1 GDIP01111288 JAL92426.1 GECL01003416 JAP02708.1 GDIP01078937 JAM24778.1 GBGD01000818 JAC88071.1 GDIQ01102147 JAL49579.1 GFJQ02007419 JAV99550.1 GDHC01009487 GDHC01004840 JAQ09142.1 JAQ13789.1 GBHO01031046 GBHO01027525 GBHO01027524 GBHO01027523 GBHO01027522 GBHO01027519 GBHO01027518 GBHO01027517 GBHO01018558 GBHO01010776 GBHO01010773 GBHO01010772 JAG12558.1 JAG16079.1 JAG16080.1 JAG16081.1 JAG16082.1 JAG16085.1 JAG16086.1 JAG16087.1 JAG25046.1 JAG32828.1 JAG32831.1 JAG32832.1 ACPB03004561 KK120151 KFM77636.1 GBRD01007034 JAG58787.1 LJIJ01001583 ODM91354.1 GDIQ01007777 JAN86960.1 GDIP01066101 JAM37614.1 GDIP01008729 JAM94986.1 CM000157 EDW88301.1 GBXI01010534 JAD03758.1 GDRN01029748 JAI67587.1 AY052131 AAK93555.1 AMQN01008724 KB303819 ELU02742.1 AE014134 BT150135 AAF53293.1 AGO63478.1 CM000361 CM002910 EDX04916.1 KMY90087.1 CH954177 EDV58689.1 CH480831 EDW46034.1 GAKP01003873 GAKP01003872 JAC55080.1 GANP01008370 JAB76098.1 GANP01008371 JAB76097.1 GDHF01014243 JAI38071.1 GDHF01029179 JAI23135.1
KQ459594 KPI96168.1 AGBW02007730 OWR54657.1 KQ461108 KPJ09218.1 ODYU01001651 SOQ38171.1 JTDY01003791 KOB69003.1 GEZM01095270 JAV55436.1 LJIG01022528 KRT80104.1 GGFK01009398 MBW42719.1 GGFK01009399 MBW42720.1 NEVH01011186 PNF32184.1 CH477499 EAT39913.1 GAPW01000479 JAC13119.1 KQ971382 EEZ97186.1 ADMH02001180 ETN63816.1 GGFJ01003213 MBW52354.1 GGFM01006050 MBW26801.1 GGFJ01003209 MBW52350.1 APCN01005234 JXUM01025946 KQ560739 KXJ81065.1 AAAB01008980 EAA14324.4 AXCN02001858 KK852470 KDR23255.1 GFDL01009390 JAV25655.1 GFDL01009314 JAV25731.1 GFDL01009267 JAV25778.1 PYGN01000002 PSN58434.1 DS233443 EDS30028.1 GFDL01009152 JAV25893.1 GFDL01009097 JAV25948.1 UFQS01000396 UFQS01001481 UFQT01000396 UFQT01001481 SSX03604.1 SSX11169.1 SSX23969.1 KB632309 ERL92070.1 APGK01044530 APGK01044531 KB741026 ENN74957.1 ENN74958.1 GFDL01009387 JAV25658.1 GACK01005077 JAA59957.1 GFDL01009469 JAV25576.1 IAAA01005327 LAA01943.1 AJVK01010273 AJVK01010274 AJVK01010275 AJVK01010276 IAAA01005328 LAA01944.1 AXCM01000229 GACK01003482 JAA61552.1 PZQS01000002 PVD35860.1 GFDL01009231 JAV25814.1 GDIP01150040 JAJ73362.1 GDIQ01137071 JAL14655.1 GDIP01217568 JAJ05834.1 GFWV01019155 MAA43883.1 AJWK01035530 GDIP01166679 GDIP01050394 LRGB01000024 JAM53321.1 KZS21530.1 ATLV01022925 KE525338 KFB48123.1 GFDL01009248 JAV25797.1 GFTR01007576 JAW08850.1 GDIQ01089501 JAN05236.1 GDIP01187928 JAJ35474.1 GFAC01003991 JAT95197.1 GDIP01111288 JAL92426.1 GECL01003416 JAP02708.1 GDIP01078937 JAM24778.1 GBGD01000818 JAC88071.1 GDIQ01102147 JAL49579.1 GFJQ02007419 JAV99550.1 GDHC01009487 GDHC01004840 JAQ09142.1 JAQ13789.1 GBHO01031046 GBHO01027525 GBHO01027524 GBHO01027523 GBHO01027522 GBHO01027519 GBHO01027518 GBHO01027517 GBHO01018558 GBHO01010776 GBHO01010773 GBHO01010772 JAG12558.1 JAG16079.1 JAG16080.1 JAG16081.1 JAG16082.1 JAG16085.1 JAG16086.1 JAG16087.1 JAG25046.1 JAG32828.1 JAG32831.1 JAG32832.1 ACPB03004561 KK120151 KFM77636.1 GBRD01007034 JAG58787.1 LJIJ01001583 ODM91354.1 GDIQ01007777 JAN86960.1 GDIP01066101 JAM37614.1 GDIP01008729 JAM94986.1 CM000157 EDW88301.1 GBXI01010534 JAD03758.1 GDRN01029748 JAI67587.1 AY052131 AAK93555.1 AMQN01008724 KB303819 ELU02742.1 AE014134 BT150135 AAF53293.1 AGO63478.1 CM000361 CM002910 EDX04916.1 KMY90087.1 CH954177 EDV58689.1 CH480831 EDW46034.1 GAKP01003873 GAKP01003872 JAC55080.1 GANP01008370 JAB76098.1 GANP01008371 JAB76097.1 GDHF01014243 JAI38071.1 GDHF01029179 JAI23135.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000235965 UP000075903 UP000069272 UP000008820 UP000075880 UP000007266 UP000000673 UP000076408 UP000075885 UP000075882 UP000075840 UP000069940 UP000249989 UP000007062 UP000075920 UP000075886 UP000027135 UP000075902 UP000245037 UP000002320 UP000076407 UP000075900 UP000030742 UP000019118 UP000092462 UP000075883 UP000245119 UP000092461 UP000076858 UP000030765 UP000075884 UP000015103 UP000054359 UP000094527 UP000002282 UP000014760 UP000000803 UP000000304 UP000008711 UP000001292
UP000235965 UP000075903 UP000069272 UP000008820 UP000075880 UP000007266 UP000000673 UP000076408 UP000075885 UP000075882 UP000075840 UP000069940 UP000249989 UP000007062 UP000075920 UP000075886 UP000027135 UP000075902 UP000245037 UP000002320 UP000076407 UP000075900 UP000030742 UP000019118 UP000092462 UP000075883 UP000245119 UP000092461 UP000076858 UP000030765 UP000075884 UP000015103 UP000054359 UP000094527 UP000002282 UP000014760 UP000000803 UP000000304 UP000008711 UP000001292
Interpro
SUPFAM
SSF52833
SSF52833
Gene 3D
ProteinModelPortal
H9ITT1
A0A2W1BSY7
A0A2A4JWB4
A0A2A4JVP2
A0A194PT73
A0A212FLP9
+ More
A0A194QUL3 A0A2H1VBI6 A0A0L7L0H7 A0A1Y1K7N1 A0A0T6AYD4 A0A2M4APL7 A0A2M4APM1 A0A2J7QUC2 A0A182VDE1 A0A182FMU9 Q16Z37 A0A182IMN5 A0A023EXG3 D6X529 W5JI60 A0A2M4BH54 A0A2M3ZE94 A0A2M4BH18 A0A182YHC3 A0A182P8D0 A0A182KJS6 A0A182I0Q8 A0A182H400 Q7PM55 A0A182VSY5 A0A182QMP7 A0A067RRK1 A0A1Q3FDK9 A0A1Q3FDU5 A0A1Q3FE39 A0A182TDT2 A0A2P8ZPK7 B0XJ65 A0A1Q3FEB5 A0A1S4H450 A0A1Q3FEJ4 A0A182XB64 A0A182R6V8 A0A336KFI3 U4UPW9 N6U8M7 N6TB44 A0A1Q3FDT6 L7M7V9 A0A1Q3FDD0 A0A2L2Y199 A0A1B0D1K1 A0A2L2Y1B4 A0A182LXR6 L7MCN0 A0A2T7PR44 A0A1Q3FE34 A0A0P5DZK6 A0A0P5NVM7 A0A0P5A1Z0 A0A293MSL5 A0A1B0CY75 A0A0P5Z6H7 A0A084WD29 A0A1Q3FE26 A0A224XK72 A0A0P6CPQ6 A0A0P5B8M0 A0A1E1X7E8 A0A0P5UUR0 A0A0V0G3Q8 A0A0P5X0Y9 A0A182N8Q5 A0A069DW52 A0A0N8C9B4 A0A1Z5KXN5 A0A146M2Y5 A0A0A9YNZ9 T1I3Q3 A0A087UJU7 A0A0K8SZU8 A0A1D2MEP7 A0A0P6I972 A0A0P5XZ73 A0A0P6BYC2 B4P359 A0A0A1WZD6 A0A0P4WKC0 Q960C0 R7UHJ2 Q9VJZ1 B4Q4S8 B3NAX5 B4IEK7 A0A034WLQ1 V5HQ91 V5HL97 A0A0K8VGQ5 A0A0K8U9W2
A0A194QUL3 A0A2H1VBI6 A0A0L7L0H7 A0A1Y1K7N1 A0A0T6AYD4 A0A2M4APL7 A0A2M4APM1 A0A2J7QUC2 A0A182VDE1 A0A182FMU9 Q16Z37 A0A182IMN5 A0A023EXG3 D6X529 W5JI60 A0A2M4BH54 A0A2M3ZE94 A0A2M4BH18 A0A182YHC3 A0A182P8D0 A0A182KJS6 A0A182I0Q8 A0A182H400 Q7PM55 A0A182VSY5 A0A182QMP7 A0A067RRK1 A0A1Q3FDK9 A0A1Q3FDU5 A0A1Q3FE39 A0A182TDT2 A0A2P8ZPK7 B0XJ65 A0A1Q3FEB5 A0A1S4H450 A0A1Q3FEJ4 A0A182XB64 A0A182R6V8 A0A336KFI3 U4UPW9 N6U8M7 N6TB44 A0A1Q3FDT6 L7M7V9 A0A1Q3FDD0 A0A2L2Y199 A0A1B0D1K1 A0A2L2Y1B4 A0A182LXR6 L7MCN0 A0A2T7PR44 A0A1Q3FE34 A0A0P5DZK6 A0A0P5NVM7 A0A0P5A1Z0 A0A293MSL5 A0A1B0CY75 A0A0P5Z6H7 A0A084WD29 A0A1Q3FE26 A0A224XK72 A0A0P6CPQ6 A0A0P5B8M0 A0A1E1X7E8 A0A0P5UUR0 A0A0V0G3Q8 A0A0P5X0Y9 A0A182N8Q5 A0A069DW52 A0A0N8C9B4 A0A1Z5KXN5 A0A146M2Y5 A0A0A9YNZ9 T1I3Q3 A0A087UJU7 A0A0K8SZU8 A0A1D2MEP7 A0A0P6I972 A0A0P5XZ73 A0A0P6BYC2 B4P359 A0A0A1WZD6 A0A0P4WKC0 Q960C0 R7UHJ2 Q9VJZ1 B4Q4S8 B3NAX5 B4IEK7 A0A034WLQ1 V5HQ91 V5HL97 A0A0K8VGQ5 A0A0K8U9W2
PDB
3IDV
E-value=9.66996e-31,
Score=335
Ontologies
GO
Topology
SignalP
Position: 1 - 24,
Likelihood: 0.949636
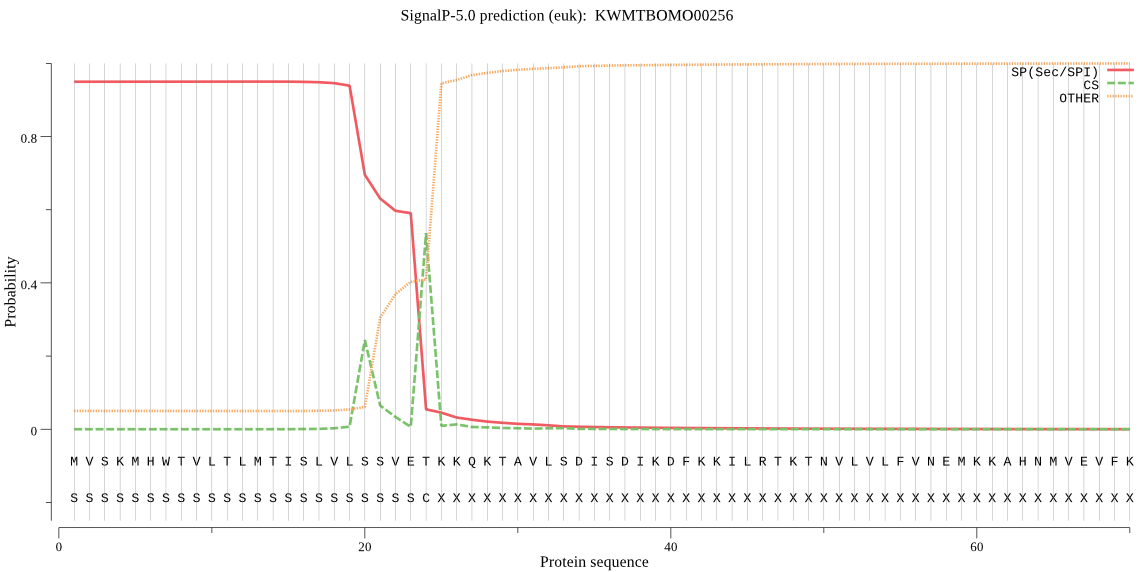
Length:
644
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.07642
Exp number, first 60 AAs:
0.85796
Total prob of N-in:
0.04641
outside
1 - 644

Population Genetic Test Statistics
Pi
7.593617
Theta
19.956325
Tajima's D
-1.569334
CLR
0.007921
CSRT
0.0515474226288686
Interpretation
Uncertain
