Gene
KWMTBOMO00254 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000559
Annotation
triosephosphate_isomerase_[Bombyx_mori]
Full name
Triosephosphate isomerase
Alternative Name
Triose-phosphate isomerase
Location in the cell
Cytoplasmic Reliability : 3.393
Sequence
CDS
ATGGGTCGCAAATTTGTTGTTGGAGGTAACTGGAAGATGAATGGTGACAAAAATCAAATCAACGAAATAGTTAATAATCTAAAAAAGGGACCTCTAGATCCTAATGTTGAGGTAATTGTTGGTGTTCCTGCTATTTACCTGTCTTATGTGAAGACTATCATTCCTGATAATGTCGAAGTTGCAGCCCAGAACTGTTGGAAGTCTCCAAAAGGAGCTTTTACTGGTGAAATTTCACCCGCTATGATCAAAGACGTTGGAGTAAATTGGGTTATACTTGGTCATTCTGAAAGAAGAACAATTTTTGGTGAAAAAGATGAACTTGTTGCTGAAAAGGTTGCTCATGCCCTTGAATCTGGCTTGAAAGTCATTGCCTGTATTGGTGAGACTCTCGAAGAGAGAGAATCTGGCAAGACTGAGGAAGTTGTTTTTAGACAATTAAAAGCTTTAGTATCGGCCATTGGTGACAAATGGGAAAATATTGTGCTTGCCTATGAACCTGTATGGGCTATTGGTACAGGCAAGACTGCTACTCCCCAACAGGCTCAAGATGTCCACCATGCTCTTCGTAACTGGTTGTCGGCCAATGTGAGTGGGAGTGTATCTGACGCTGTACGTATTCAGTACGGTGGCTCGGTCACAGCTGCTAATGCTAAGGAGCTTGCATCCTGCAAAGACATTGATGGTTTCTTAGTTGGTGGAGCCAGTCTTAAGCCTGAGTTTGTAGAAATTGTAAATGCTAATCAATAA
Protein
MGRKFVVGGNWKMNGDKNQINEIVNNLKKGPLDPNVEVIVGVPAIYLSYVKTIIPDNVEVAAQNCWKSPKGAFTGEISPAMIKDVGVNWVILGHSERRTIFGEKDELVAEKVAHALESGLKVIACIGETLEERESGKTEEVVFRQLKALVSAIGDKWENIVLAYEPVWAIGTGKTATPQQAQDVHHALRNWLSANVSGSVSDAVRIQYGGSVTAANAKELASCKDIDGFLVGGASLKPEFVEIVNANQ
Summary
Catalytic Activity
D-glyceraldehyde 3-phosphate = dihydroxyacetone phosphate
Subunit
Homodimer.
Similarity
Belongs to the triosephosphate isomerase family.
Keywords
Complete proteome
Direct protein sequencing
Gluconeogenesis
Glycolysis
Isomerase
Reference proteome
Feature
chain Triosephosphate isomerase
Uniprot
P82204
A0A194PS77
A0A2W1BMP5
Q5XUN5
A0A2H1VCN8
A0A2A4JXG2
+ More
S4P3S7 A0A212FLN4 E3UKQ5 D2A042 A0A1Y1NGV6 A0A0L7KX23 Q8MPF2 A0A232EQN8 K7J7M3 J3JWZ9 U4U647 A4ZXC4 N6UJG6 A0A026WB62 I6P143 V9IKD3 A0A1L1ZLV2 A0A084VWK0 A0A182PGM5 A0A0K8TPC5 A0A182FQ32 A0A2M4BXU2 A0A182UTK0 A0A182UA61 A0A182L9S9 Q7PXW5 A0A182MMU2 A0A182YB73 A0A182IWG6 A0A182WSJ2 A0A182HLE4 A0A2M3Z499 A0A2M3ZGK0 A0A2M4A9W5 A0A195DCJ8 A0A182QLI5 V5SJ02 A0A151IGF3 A0A0C9QY89 A0A182VZD4 T1DMV1 A0A182K2M8 U5EUW5 A0A182R608 A0A182NS29 A0A151WFE9 E2AJK8 F4WM74 A0A182SBQ1 A0A1B0ETY1 A0A1I9WLF0 W5J8Q9 E2BX54 A0A0J7KLG9 A0A1I9VT67 A0A1B6LPQ3 A0A154PA23 A0A158P300 A0A195F2F0 E0VJK8 A0A0L7RCW0 Q09KA2 A0A1L8E289 A0A1B6FCS1 A0A2P0XIH0 A0A293MVI9 A0A1B6DQP8 Q17HW3 A0A1B6BWW9 L7UZA7 P30741 A0A1L8E1V1 P55275 A0A2P8XY09 A0A182H0S2 A0A023END4 C4WT45 W8BWJ7 A0A1Z5KWQ3 A0A0C5Q4W3 A2I3Y4 A0A2H8TXV9 A0A3L8DRN9 W8C8K9 T1DII6 A0A0P9ATG2 A0A0K8RFH3 A0A131Y250 A0A0A1WUG1 A0A0A1XKP0 R4UMY9 A0A0P5Z6D0 A0A023FUT8 A0A336LFY4
S4P3S7 A0A212FLN4 E3UKQ5 D2A042 A0A1Y1NGV6 A0A0L7KX23 Q8MPF2 A0A232EQN8 K7J7M3 J3JWZ9 U4U647 A4ZXC4 N6UJG6 A0A026WB62 I6P143 V9IKD3 A0A1L1ZLV2 A0A084VWK0 A0A182PGM5 A0A0K8TPC5 A0A182FQ32 A0A2M4BXU2 A0A182UTK0 A0A182UA61 A0A182L9S9 Q7PXW5 A0A182MMU2 A0A182YB73 A0A182IWG6 A0A182WSJ2 A0A182HLE4 A0A2M3Z499 A0A2M3ZGK0 A0A2M4A9W5 A0A195DCJ8 A0A182QLI5 V5SJ02 A0A151IGF3 A0A0C9QY89 A0A182VZD4 T1DMV1 A0A182K2M8 U5EUW5 A0A182R608 A0A182NS29 A0A151WFE9 E2AJK8 F4WM74 A0A182SBQ1 A0A1B0ETY1 A0A1I9WLF0 W5J8Q9 E2BX54 A0A0J7KLG9 A0A1I9VT67 A0A1B6LPQ3 A0A154PA23 A0A158P300 A0A195F2F0 E0VJK8 A0A0L7RCW0 Q09KA2 A0A1L8E289 A0A1B6FCS1 A0A2P0XIH0 A0A293MVI9 A0A1B6DQP8 Q17HW3 A0A1B6BWW9 L7UZA7 P30741 A0A1L8E1V1 P55275 A0A2P8XY09 A0A182H0S2 A0A023END4 C4WT45 W8BWJ7 A0A1Z5KWQ3 A0A0C5Q4W3 A2I3Y4 A0A2H8TXV9 A0A3L8DRN9 W8C8K9 T1DII6 A0A0P9ATG2 A0A0K8RFH3 A0A131Y250 A0A0A1WUG1 A0A0A1XKP0 R4UMY9 A0A0P5Z6D0 A0A023FUT8 A0A336LFY4
EC Number
5.3.1.1
Pubmed
11280994
26354079
28756777
23622113
22118469
18362917
+ More
19820115 28004739 26227816 19849721 28648823 20075255 22516182 23537049 24508170 23569222 24438588 26369729 20966253 12364791 14747013 17210077 25244985 20798317 21719571 27538518 20920257 23761445 21347285 20566863 17510324 8429888 7667320 29403074 26483478 24945155 24495485 28528879 25702953 30249741 24330624 17994087 25830018
19820115 28004739 26227816 19849721 28648823 20075255 22516182 23537049 24508170 23569222 24438588 26369729 20966253 12364791 14747013 17210077 25244985 20798317 21719571 27538518 20920257 23761445 21347285 20566863 17510324 8429888 7667320 29403074 26483478 24945155 24495485 28528879 25702953 30249741 24330624 17994087 25830018
EMBL
AY734490
AAU34185.1
KQ459594
KPI96167.1
KZ149961
PZC76329.1
+ More
AY736358 AAU84716.2 ODYU01001651 SOQ38172.1 NWSH01000478 PCG76110.1 GAIX01011480 JAA81080.1 AGBW02007730 OWR54655.1 HM449929 ADO33064.1 KQ971338 EFA01748.1 GEZM01002999 JAV96999.1 JTDY01004902 KOB67611.1 AJ496728 CAD43178.1 NNAY01002764 OXU20616.1 AAZX01016602 BT127767 AEE62729.1 KB632166 ERL89334.1 EF493864 EU760983 ABP65286.1 ACE95726.1 APGK01032042 KB740834 ENN78802.1 KK107293 EZA53337.1 JQ771343 AFD93374.1 JR048821 AEY60774.1 KP994676 KP994677 ALG65074.1 ATLV01017623 KE525179 KFB42344.1 GDAI01001607 JAI15996.1 GGFJ01008755 MBW57896.1 AAAB01008987 EAA00928.4 AXCM01001257 APCN01000895 GGFM01002596 MBW23347.1 GGFM01006901 MBW27652.1 GGFK01004234 MBW37555.1 KQ980989 KYN10562.1 AXCN02000266 KF647619 AHB50481.1 KQ977726 KYN00292.1 GBYB01005647 JAG75414.1 GAMD01003425 JAA98165.1 GANO01001272 JAB58599.1 KQ983219 KYQ46533.1 GL440027 EFN66403.1 GL888217 EGI64718.1 AJWK01013578 KU932334 APA33970.1 ADMH02002100 ETN59184.1 GL451202 EFN79773.1 LBMM01005731 KMQ91258.1 KX024698 APA16886.1 GEBQ01014380 JAT25597.1 KQ434856 KZC08702.1 ADTU01001303 KQ981855 KYN34755.1 DS235225 EEB13564.1 KQ414614 KOC68827.1 DQ885469 ABI63547.1 GFDF01001459 JAV12625.1 GECZ01021767 JAS48002.1 KY661336 AVA17423.1 GFWV01019230 MAA43958.1 GEDC01009301 JAS27997.1 CH477245 EAT46226.1 GEDC01031516 JAS05782.1 KC305500 AGC56216.1 L07390 GFDF01001458 JAV12626.1 U23080 PYGN01001178 PSN36887.1 JXUM01022746 KQ560640 KXJ81473.1 JXUM01021307 GAPW01003704 KQ560596 JAC09894.1 KXJ81709.1 ABLF02029934 AK340461 BAH71065.1 GAMC01002963 JAC03593.1 GFJQ02007418 JAV99551.1 KM609492 AJQ30116.1 EF070487 ABM55553.1 GFXV01006935 MBW18740.1 QOIP01000005 RLU23074.1 GAMC01002964 JAC03592.1 GALA01000911 JAA93941.1 CH902617 KPU80615.1 GADI01003891 JAA69917.1 GEFM01002839 JAP72957.1 GBXI01012137 JAD02155.1 GBXI01016352 GBXI01003174 JAC97939.1 JAD11118.1 KC571986 AGM32485.1 GDIP01048324 LRGB01002849 JAM55391.1 KZS06051.1 GBBL01001744 JAC25576.1 UFQS01003368 UFQT01003368 SSX15561.1 SSX34925.1
AY736358 AAU84716.2 ODYU01001651 SOQ38172.1 NWSH01000478 PCG76110.1 GAIX01011480 JAA81080.1 AGBW02007730 OWR54655.1 HM449929 ADO33064.1 KQ971338 EFA01748.1 GEZM01002999 JAV96999.1 JTDY01004902 KOB67611.1 AJ496728 CAD43178.1 NNAY01002764 OXU20616.1 AAZX01016602 BT127767 AEE62729.1 KB632166 ERL89334.1 EF493864 EU760983 ABP65286.1 ACE95726.1 APGK01032042 KB740834 ENN78802.1 KK107293 EZA53337.1 JQ771343 AFD93374.1 JR048821 AEY60774.1 KP994676 KP994677 ALG65074.1 ATLV01017623 KE525179 KFB42344.1 GDAI01001607 JAI15996.1 GGFJ01008755 MBW57896.1 AAAB01008987 EAA00928.4 AXCM01001257 APCN01000895 GGFM01002596 MBW23347.1 GGFM01006901 MBW27652.1 GGFK01004234 MBW37555.1 KQ980989 KYN10562.1 AXCN02000266 KF647619 AHB50481.1 KQ977726 KYN00292.1 GBYB01005647 JAG75414.1 GAMD01003425 JAA98165.1 GANO01001272 JAB58599.1 KQ983219 KYQ46533.1 GL440027 EFN66403.1 GL888217 EGI64718.1 AJWK01013578 KU932334 APA33970.1 ADMH02002100 ETN59184.1 GL451202 EFN79773.1 LBMM01005731 KMQ91258.1 KX024698 APA16886.1 GEBQ01014380 JAT25597.1 KQ434856 KZC08702.1 ADTU01001303 KQ981855 KYN34755.1 DS235225 EEB13564.1 KQ414614 KOC68827.1 DQ885469 ABI63547.1 GFDF01001459 JAV12625.1 GECZ01021767 JAS48002.1 KY661336 AVA17423.1 GFWV01019230 MAA43958.1 GEDC01009301 JAS27997.1 CH477245 EAT46226.1 GEDC01031516 JAS05782.1 KC305500 AGC56216.1 L07390 GFDF01001458 JAV12626.1 U23080 PYGN01001178 PSN36887.1 JXUM01022746 KQ560640 KXJ81473.1 JXUM01021307 GAPW01003704 KQ560596 JAC09894.1 KXJ81709.1 ABLF02029934 AK340461 BAH71065.1 GAMC01002963 JAC03593.1 GFJQ02007418 JAV99551.1 KM609492 AJQ30116.1 EF070487 ABM55553.1 GFXV01006935 MBW18740.1 QOIP01000005 RLU23074.1 GAMC01002964 JAC03592.1 GALA01000911 JAA93941.1 CH902617 KPU80615.1 GADI01003891 JAA69917.1 GEFM01002839 JAP72957.1 GBXI01012137 JAD02155.1 GBXI01016352 GBXI01003174 JAC97939.1 JAD11118.1 KC571986 AGM32485.1 GDIP01048324 LRGB01002849 JAM55391.1 KZS06051.1 GBBL01001744 JAC25576.1 UFQS01003368 UFQT01003368 SSX15561.1 SSX34925.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000007266
UP000037510
+ More
UP000215335 UP000002358 UP000030742 UP000019118 UP000053097 UP000030765 UP000075885 UP000069272 UP000075903 UP000075902 UP000075882 UP000007062 UP000075883 UP000076408 UP000075880 UP000076407 UP000075840 UP000078492 UP000075886 UP000078542 UP000075920 UP000075881 UP000075900 UP000075884 UP000075809 UP000000311 UP000007755 UP000075901 UP000092461 UP000000673 UP000008237 UP000036403 UP000076502 UP000005205 UP000078541 UP000009046 UP000053825 UP000008820 UP000245037 UP000069940 UP000249989 UP000007819 UP000279307 UP000007801 UP000076858
UP000215335 UP000002358 UP000030742 UP000019118 UP000053097 UP000030765 UP000075885 UP000069272 UP000075903 UP000075902 UP000075882 UP000007062 UP000075883 UP000076408 UP000075880 UP000076407 UP000075840 UP000078492 UP000075886 UP000078542 UP000075920 UP000075881 UP000075900 UP000075884 UP000075809 UP000000311 UP000007755 UP000075901 UP000092461 UP000000673 UP000008237 UP000036403 UP000076502 UP000005205 UP000078541 UP000009046 UP000053825 UP000008820 UP000245037 UP000069940 UP000249989 UP000007819 UP000279307 UP000007801 UP000076858
Pfam
PF00121 TIM
Interpro
SUPFAM
SSF51351
SSF51351
Gene 3D
CDD
ProteinModelPortal
P82204
A0A194PS77
A0A2W1BMP5
Q5XUN5
A0A2H1VCN8
A0A2A4JXG2
+ More
S4P3S7 A0A212FLN4 E3UKQ5 D2A042 A0A1Y1NGV6 A0A0L7KX23 Q8MPF2 A0A232EQN8 K7J7M3 J3JWZ9 U4U647 A4ZXC4 N6UJG6 A0A026WB62 I6P143 V9IKD3 A0A1L1ZLV2 A0A084VWK0 A0A182PGM5 A0A0K8TPC5 A0A182FQ32 A0A2M4BXU2 A0A182UTK0 A0A182UA61 A0A182L9S9 Q7PXW5 A0A182MMU2 A0A182YB73 A0A182IWG6 A0A182WSJ2 A0A182HLE4 A0A2M3Z499 A0A2M3ZGK0 A0A2M4A9W5 A0A195DCJ8 A0A182QLI5 V5SJ02 A0A151IGF3 A0A0C9QY89 A0A182VZD4 T1DMV1 A0A182K2M8 U5EUW5 A0A182R608 A0A182NS29 A0A151WFE9 E2AJK8 F4WM74 A0A182SBQ1 A0A1B0ETY1 A0A1I9WLF0 W5J8Q9 E2BX54 A0A0J7KLG9 A0A1I9VT67 A0A1B6LPQ3 A0A154PA23 A0A158P300 A0A195F2F0 E0VJK8 A0A0L7RCW0 Q09KA2 A0A1L8E289 A0A1B6FCS1 A0A2P0XIH0 A0A293MVI9 A0A1B6DQP8 Q17HW3 A0A1B6BWW9 L7UZA7 P30741 A0A1L8E1V1 P55275 A0A2P8XY09 A0A182H0S2 A0A023END4 C4WT45 W8BWJ7 A0A1Z5KWQ3 A0A0C5Q4W3 A2I3Y4 A0A2H8TXV9 A0A3L8DRN9 W8C8K9 T1DII6 A0A0P9ATG2 A0A0K8RFH3 A0A131Y250 A0A0A1WUG1 A0A0A1XKP0 R4UMY9 A0A0P5Z6D0 A0A023FUT8 A0A336LFY4
S4P3S7 A0A212FLN4 E3UKQ5 D2A042 A0A1Y1NGV6 A0A0L7KX23 Q8MPF2 A0A232EQN8 K7J7M3 J3JWZ9 U4U647 A4ZXC4 N6UJG6 A0A026WB62 I6P143 V9IKD3 A0A1L1ZLV2 A0A084VWK0 A0A182PGM5 A0A0K8TPC5 A0A182FQ32 A0A2M4BXU2 A0A182UTK0 A0A182UA61 A0A182L9S9 Q7PXW5 A0A182MMU2 A0A182YB73 A0A182IWG6 A0A182WSJ2 A0A182HLE4 A0A2M3Z499 A0A2M3ZGK0 A0A2M4A9W5 A0A195DCJ8 A0A182QLI5 V5SJ02 A0A151IGF3 A0A0C9QY89 A0A182VZD4 T1DMV1 A0A182K2M8 U5EUW5 A0A182R608 A0A182NS29 A0A151WFE9 E2AJK8 F4WM74 A0A182SBQ1 A0A1B0ETY1 A0A1I9WLF0 W5J8Q9 E2BX54 A0A0J7KLG9 A0A1I9VT67 A0A1B6LPQ3 A0A154PA23 A0A158P300 A0A195F2F0 E0VJK8 A0A0L7RCW0 Q09KA2 A0A1L8E289 A0A1B6FCS1 A0A2P0XIH0 A0A293MVI9 A0A1B6DQP8 Q17HW3 A0A1B6BWW9 L7UZA7 P30741 A0A1L8E1V1 P55275 A0A2P8XY09 A0A182H0S2 A0A023END4 C4WT45 W8BWJ7 A0A1Z5KWQ3 A0A0C5Q4W3 A2I3Y4 A0A2H8TXV9 A0A3L8DRN9 W8C8K9 T1DII6 A0A0P9ATG2 A0A0K8RFH3 A0A131Y250 A0A0A1WUG1 A0A0A1XKP0 R4UMY9 A0A0P5Z6D0 A0A023FUT8 A0A336LFY4
PDB
2I9E
E-value=6.77959e-106,
Score=979
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00562 Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
00051 Fructose and mannose metabolism - Bombyx mori (domestic silkworm)
00562 Inositol phosphate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
GO
PANTHER
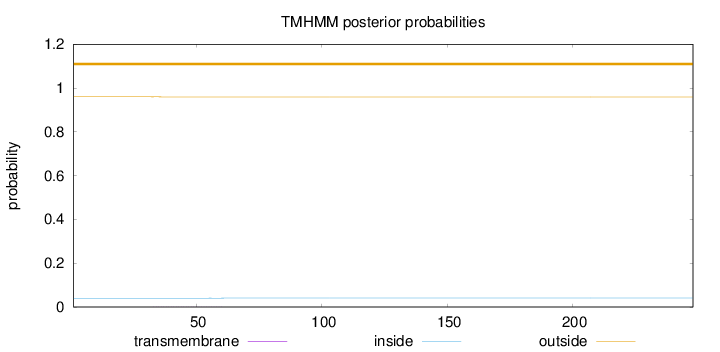
Topology
Length:
248
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02389
Exp number, first 60 AAs:
0.02168
Total prob of N-in:
0.04003
outside
1 - 248
Population Genetic Test Statistics
Pi
6.645785
Theta
12.601136
Tajima's D
-1.304235
CLR
0.053457
CSRT
0.0825958702064897
Interpretation
Uncertain
