Gene
KWMTBOMO00244 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000563
Annotation
tyrosine_hydroxylase_[Bombyx_mori]
Full name
Tyrosine 3-monooxygenase
Alternative Name
Protein Pale
Tyrosine 3-hydroxylase
Tyrosine 3-hydroxylase
Location in the cell
Cytoplasmic Reliability : 1.911 Nuclear Reliability : 2.406
Sequence
CDS
ATGGCAGTCGCAGCAGCCCAGAAGAACCGCGAAATGTTCGCCATCAAGAAATCCTACAGTATCGAGAACGGTTACCCATCTCGCCGTCGTTCCTTGGTTGATGACGCCCGCTTCGAAACACTGGTAGTCAAACAAACCAAGCAGAGCGTACTTGAAGAGGCTCGCGCCCGAGCCAATGACTCTGGCTTGGAATCTGATTTCATCCAAGATGGAGTGCATATTGGTAACGGTGATAATTCTCCAACCGTTGAAGATGGGACCCAGCAAGATGAAACGAAAAATGGTCACCTCGCTGATGCTGATATAGGCGATGACGATGCGAAGGGTGACGAGGATTATACGCTGACCGAAGAAGAAGTGATCTTGCAAAATGCTGCTAGTGAATCTTCTGAAGCGGAACAAGCTCTCCAACAAGCTGCCCTTTTATTGCATATGCGCGATGGAATGGGATCCCTCGCACGCATTCTGAAAACCATCGACAATTATAAAGGATGCGTTCAGCACCTTGAGACTCGCCCTTCCCAAGTCACTGGAGTCCAATTCGATGCTCTAGTTAAAGTCAGCATGACTCGTATTAATCTTATCCAGCTAATTAGATCCTTGCGTCAGTCAACCGCGTTTGCTGGTGTTAACCTTCTGACCGAAAATAACATCTCGAGCAAGACACCTTGGTTCCCGCGCCACGCTTCTGATCTCGATAACTGTAACCATCTCATGACCAAGTACGAACCCGAATTAGACATGAACCATCCTGGTTTCGCGGACAAAGATTACAGGGAACGCAGAAAACAGATTGCGGAAATCGCTTTCGCTTACAAATACGGTGACCCGATTCCGTCTATCGCATACACTGAAATCGAGAATGGCACTTGGCAACGAGTGTTCAACACCGTGCTTGATTTGATGCCCAAACACGCGTGCAGAGAGTACAAGGCTGCGTTCGAGAAACTACAGGCAGCAGACATATTCGTACCTCATCGTATTCCACAACTGGAAGATGTCAGTAGCTTCTTGCGTAAGCACACTGGCTTTACCCTGCGACCGGCTGCTGGTCTTCTGACGGCACGTGACTTCCTCGCCTCGCTTGCCTTCCGCGTATTCCAATCGACACAATACGTACGCCACAACAATTCGCCTTTCCACACACCTGAACCTGACTGTATTCACGAGCTTCTTGGTCATATACCATTATTGGCTGACCCCAGCTTCGCCCAATTTTCGCAAGAAATTGGCCTTGCATCCCTCGGCGCTTCGGATTCTGAAATCGAAAAACTATCTACTGTCTACTGGTTTACGGTTGAGTTCGGACTGTGCAAAGAGAACCAGCAACTGAAAGCTTATGGTGCAGCTCTTTTGTCTTCGATCGGAGAACTGCTTCATGCTCTTAGTGACAAGCCTGAGCTGAGACCGTTCGAGCCTGCATCCACATCCGTGCAACCCTACCAAGACCAGGAGTACCAGCCCATCTATTACGTAGCTGAGACCTTCGAAGATGCCAAAGACAAATTCAGACGCTGGGTGTCAACCATGTCGCGACCGTTCGAAGTGCGCTTCAACCCTCACACAGAGCGCGTAGAGGTGCTCGACTCCGTTGATAAGCTGGAAACTCTCATCTGGCAGCTCAACACCGAGATGTTGCATCTCACCAACGCCGTTAAGAAGCTCAAGGGCTCACACTTCGAGTGA
Protein
MAVAAAQKNREMFAIKKSYSIENGYPSRRRSLVDDARFETLVVKQTKQSVLEEARARANDSGLESDFIQDGVHIGNGDNSPTVEDGTQQDETKNGHLADADIGDDDAKGDEDYTLTEEEVILQNAASESSEAEQALQQAALLLHMRDGMGSLARILKTIDNYKGCVQHLETRPSQVTGVQFDALVKVSMTRINLIQLIRSLRQSTAFAGVNLLTENNISSKTPWFPRHASDLDNCNHLMTKYEPELDMNHPGFADKDYRERRKQIAEIAFAYKYGDPIPSIAYTEIENGTWQRVFNTVLDLMPKHACREYKAAFEKLQAADIFVPHRIPQLEDVSSFLRKHTGFTLRPAAGLLTARDFLASLAFRVFQSTQYVRHNNSPFHTPEPDCIHELLGHIPLLADPSFAQFSQEIGLASLGASDSEIEKLSTVYWFTVEFGLCKENQQLKAYGAALLSSIGELLHALSDKPELRPFEPASTSVQPYQDQEYQPIYYVAETFEDAKDKFRRWVSTMSRPFEVRFNPHTERVEVLDSVDKLETLIWQLNTEMLHLTNAVKKLKGSHFE
Summary
Catalytic Activity
(6R)-L-erythro-5,6,7,8-tetrahydrobiopterin + L-tyrosine + O2 = (4aS,6R)-4a-hydroxy-L-erythro-5,6,7,8-tetrahydrobiopterin + L-dopa
Cofactor
Fe(2+)
Similarity
Belongs to the biopterin-dependent aromatic amino acid hydroxylase family.
Keywords
Alternative splicing
Catecholamine biosynthesis
Complete proteome
Iron
Metal-binding
Monooxygenase
Neurotransmitter biosynthesis
Oxidoreductase
Reference proteome
Feature
chain Tyrosine 3-monooxygenase
splice variant In isoform Hypodermal.
splice variant In isoform Hypodermal.
Uniprot
B7XAK2
E9L3G5
E9L3H1
E9L3J0
E9L3H7
E9L3J1
+ More
E9L3J2 E9L3G7 E9L3G4 E9L3H0 E9L3H5 E9L3H6 E9L3H8 E9L3I0 E7CZD7 A0A2H1VJX9 A5YVV1 A0A2W1BU82 M4GVG2 A0A2A4K6A9 A0A0G3VI81 Q402L8 D5MRL1 Q0E9S3 A0A194PT63 D5MRK5 D5LPT9 A6BLY5 F2VLL9 A0A212FDY4 D9IX57 A0A0L7L763 T1TCT2 A0A0G3VFY8 Q0E9S2 A0A182QXT4 A0A182WKJ2 A0A182RPF1 A0A182YQC3 A0A182V967 A0A182KPE6 A0A182X777 Q7PII7 A0A182I1Q6 A0A195BCD3 E2BF32 T1DPG9 Q29DR7 B4N2X5 B4H7A4 A0A182IZD0 A0A026WHN9 A0A3B0KGP5 A0A158NFE6 A0A1W4UXG4 E9IJ93 A0A182PJJ2 A0A0L0CQQ4 B4J096 A0A182FDH1 B4KW72 A0A084W0Q1 F4WGX8 A0A1Q3FU03 A5YVV2 A0A154P317 B4LHH8 B5X0J4 P18459 A0A1A9XWL5 F8J2H7 A0A1B0B587 B3M9H5 A0A336LN07 A0A1B0FBP0 E2A6B6 B3NFM6 A0A1A9Z3A0 A0A1A9VYP6 A0A0J9RQ65 B4PJR3 A0A2P8YCA1 B4QJT1 A0A0C9RB22 T1PBG7 G8E099 A0A1Y1M337 W5JBY1 A0A0Q9X3P9 W8BHQ1 A0A0A9WVH7 A0A034VJ95 A0A2R8G1S9 A0A0A1XT79 A0A0J7L2G0 A0A1W4VAQ6 A0A0K8VQZ7 D6WHA4 A0A1L8E390 C8CE48
E9L3J2 E9L3G7 E9L3G4 E9L3H0 E9L3H5 E9L3H6 E9L3H8 E9L3I0 E7CZD7 A0A2H1VJX9 A5YVV1 A0A2W1BU82 M4GVG2 A0A2A4K6A9 A0A0G3VI81 Q402L8 D5MRL1 Q0E9S3 A0A194PT63 D5MRK5 D5LPT9 A6BLY5 F2VLL9 A0A212FDY4 D9IX57 A0A0L7L763 T1TCT2 A0A0G3VFY8 Q0E9S2 A0A182QXT4 A0A182WKJ2 A0A182RPF1 A0A182YQC3 A0A182V967 A0A182KPE6 A0A182X777 Q7PII7 A0A182I1Q6 A0A195BCD3 E2BF32 T1DPG9 Q29DR7 B4N2X5 B4H7A4 A0A182IZD0 A0A026WHN9 A0A3B0KGP5 A0A158NFE6 A0A1W4UXG4 E9IJ93 A0A182PJJ2 A0A0L0CQQ4 B4J096 A0A182FDH1 B4KW72 A0A084W0Q1 F4WGX8 A0A1Q3FU03 A5YVV2 A0A154P317 B4LHH8 B5X0J4 P18459 A0A1A9XWL5 F8J2H7 A0A1B0B587 B3M9H5 A0A336LN07 A0A1B0FBP0 E2A6B6 B3NFM6 A0A1A9Z3A0 A0A1A9VYP6 A0A0J9RQ65 B4PJR3 A0A2P8YCA1 B4QJT1 A0A0C9RB22 T1PBG7 G8E099 A0A1Y1M337 W5JBY1 A0A0Q9X3P9 W8BHQ1 A0A0A9WVH7 A0A034VJ95 A0A2R8G1S9 A0A0A1XT79 A0A0J7L2G0 A0A1W4VAQ6 A0A0K8VQZ7 D6WHA4 A0A1L8E390 C8CE48
EC Number
1.14.16.2
Pubmed
18854583
19121390
21212153
17967351
28756777
16133568
+ More
26354079 18178496 22118469 26227816 25244985 20966253 12364791 14747013 17210077 20798317 15632085 17994087 24508170 30249741 21347285 21282665 26108605 24438588 27416870 21719571 20080183 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 2483109 12537569 22936249 17550304 29403074 25315136 28004739 20920257 23761445 24495485 25401762 26823975 25348373 28941310 25830018 18362917 19820115
26354079 18178496 22118469 26227816 25244985 20966253 12364791 14747013 17210077 20798317 15632085 17994087 24508170 30249741 21347285 21282665 26108605 24438588 27416870 21719571 20080183 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 2483109 12537569 22936249 17550304 29403074 25315136 28004739 20920257 23761445 24495485 25401762 26823975 25348373 28941310 25830018 18362917 19820115
EMBL
BABH01006925
GQ387061
HM545522
HM545524
HM545525
HM545526
+ More
HM545527 HM545528 HM545529 HM545530 HM545531 HM545532 AB439286 ACU82844.1 ADV56717.1 ADV56719.1 ADV56720.1 ADV56721.1 ADV56722.1 ADV56723.1 ADV56724.1 ADV56725.1 ADV56726.1 ADV56727.1 BAH11148.1 HM545508 HM545509 HM545511 HM545512 HM545515 HM545516 HM545517 HM545536 ADV56703.1 ADV56704.1 ADV56706.1 ADV56707.1 ADV56710.1 ADV56711.1 ADV56712.1 ADV56731.1 HM545514 ADV56709.1 HM545533 ADV56728.1 HM545520 ADV56715.1 HM545534 ADV56729.1 HM545535 ADV56730.1 HM545510 ADV56705.1 HM545507 ADV56702.1 HM545513 ADV56708.1 HM545518 ADV56713.1 HM545519 ADV56714.1 HM545521 ADV56716.1 HM545523 ADV56718.1 HM856623 ADV32917.1 ODYU01002965 SOQ41135.1 EF592177 ABQ95973.1 MF440319 KZ149961 ASS36971.1 PZC76340.1 JF795467 JF795468 AFG25778.1 AFG25779.1 NWSH01000125 PCG79303.1 KP657623 RSAL01000039 AKL78848.1 RVE51020.1 AB178006 BAE43824.1 AB525743 BAJ07593.1 AB274834 BAF32573.1 KQ459594 KPI96158.1 AB525737 KQ461108 BAJ07587.1 KPJ09226.1 GU953229 ADF43213.1 AB288228 BAF64534.1 GU386339 ADU32895.1 AGBW02008997 OWR51918.1 HM215247 JN410829 ADK12633.1 AEP25405.1 JTDY01002569 KOB71174.1 KC996753 AGT62463.1 KP657624 AKL78849.1 AB274835 BAF32574.1 AXCN02000407 AAAB01008960 EAA44132.3 APCN01000085 KQ976514 KYM82221.1 GL447910 EFN85710.1 GAMD01003344 JAA98246.1 CH379070 EAL30347.2 CH964062 EDW78714.1 CH479217 EDW33710.1 KK107199 QOIP01000009 EZA55545.1 RLU18977.1 OUUW01000012 SPP87590.1 ADTU01014360 GL763739 EFZ19360.1 JRES01000036 KNC34688.1 CH916366 EDV95697.1 CH933809 EDW17460.1 ATLV01019149 KU886220 KE525263 AMZ03511.1 KFB43795.1 GL888147 EGI66597.1 GFDL01003971 JAV31074.1 EF592178 ABQ95974.1 KQ434795 KZC05530.1 CH940647 EDW68508.1 BT044563 AE014296 ACI15758.1 AHN57980.1 U14395 X76209 AY071698 FJ830627 ACZ06878.1 JXJN01008642 JXJN01008643 CH902618 EDV41188.1 UFQS01000067 UFQT01000067 SSW98948.1 SSX19330.1 CCAG010001557 GL437123 EFN71001.1 CH954178 EDV50568.1 CM002912 KMY97958.1 CM000159 EDW93662.1 PYGN01000708 PSN41889.1 CM000363 EDX09472.1 GBYB01013689 JAG83456.1 KA646004 AFP60633.1 JF499654 AEC14314.1 GEZM01042393 JAV79891.1 ADMH02001663 ETN61516.1 KRF98935.1 GAMC01017321 JAB89234.1 GBHO01032173 GBRD01000651 GDHC01016939 JAG11431.1 JAG65170.1 JAQ01690.1 GAKP01016775 JAC42177.1 KY911196 AVP73886.1 GBXI01000132 JAD14160.1 LBMM01001144 KMQ96828.1 GDHF01011012 JAI41302.1 KQ971331 EEZ99729.1 GFDF01001119 JAV12965.1 GQ390366 ACU77882.2
HM545527 HM545528 HM545529 HM545530 HM545531 HM545532 AB439286 ACU82844.1 ADV56717.1 ADV56719.1 ADV56720.1 ADV56721.1 ADV56722.1 ADV56723.1 ADV56724.1 ADV56725.1 ADV56726.1 ADV56727.1 BAH11148.1 HM545508 HM545509 HM545511 HM545512 HM545515 HM545516 HM545517 HM545536 ADV56703.1 ADV56704.1 ADV56706.1 ADV56707.1 ADV56710.1 ADV56711.1 ADV56712.1 ADV56731.1 HM545514 ADV56709.1 HM545533 ADV56728.1 HM545520 ADV56715.1 HM545534 ADV56729.1 HM545535 ADV56730.1 HM545510 ADV56705.1 HM545507 ADV56702.1 HM545513 ADV56708.1 HM545518 ADV56713.1 HM545519 ADV56714.1 HM545521 ADV56716.1 HM545523 ADV56718.1 HM856623 ADV32917.1 ODYU01002965 SOQ41135.1 EF592177 ABQ95973.1 MF440319 KZ149961 ASS36971.1 PZC76340.1 JF795467 JF795468 AFG25778.1 AFG25779.1 NWSH01000125 PCG79303.1 KP657623 RSAL01000039 AKL78848.1 RVE51020.1 AB178006 BAE43824.1 AB525743 BAJ07593.1 AB274834 BAF32573.1 KQ459594 KPI96158.1 AB525737 KQ461108 BAJ07587.1 KPJ09226.1 GU953229 ADF43213.1 AB288228 BAF64534.1 GU386339 ADU32895.1 AGBW02008997 OWR51918.1 HM215247 JN410829 ADK12633.1 AEP25405.1 JTDY01002569 KOB71174.1 KC996753 AGT62463.1 KP657624 AKL78849.1 AB274835 BAF32574.1 AXCN02000407 AAAB01008960 EAA44132.3 APCN01000085 KQ976514 KYM82221.1 GL447910 EFN85710.1 GAMD01003344 JAA98246.1 CH379070 EAL30347.2 CH964062 EDW78714.1 CH479217 EDW33710.1 KK107199 QOIP01000009 EZA55545.1 RLU18977.1 OUUW01000012 SPP87590.1 ADTU01014360 GL763739 EFZ19360.1 JRES01000036 KNC34688.1 CH916366 EDV95697.1 CH933809 EDW17460.1 ATLV01019149 KU886220 KE525263 AMZ03511.1 KFB43795.1 GL888147 EGI66597.1 GFDL01003971 JAV31074.1 EF592178 ABQ95974.1 KQ434795 KZC05530.1 CH940647 EDW68508.1 BT044563 AE014296 ACI15758.1 AHN57980.1 U14395 X76209 AY071698 FJ830627 ACZ06878.1 JXJN01008642 JXJN01008643 CH902618 EDV41188.1 UFQS01000067 UFQT01000067 SSW98948.1 SSX19330.1 CCAG010001557 GL437123 EFN71001.1 CH954178 EDV50568.1 CM002912 KMY97958.1 CM000159 EDW93662.1 PYGN01000708 PSN41889.1 CM000363 EDX09472.1 GBYB01013689 JAG83456.1 KA646004 AFP60633.1 JF499654 AEC14314.1 GEZM01042393 JAV79891.1 ADMH02001663 ETN61516.1 KRF98935.1 GAMC01017321 JAB89234.1 GBHO01032173 GBRD01000651 GDHC01016939 JAG11431.1 JAG65170.1 JAQ01690.1 GAKP01016775 JAC42177.1 KY911196 AVP73886.1 GBXI01000132 JAD14160.1 LBMM01001144 KMQ96828.1 GDHF01011012 JAI41302.1 KQ971331 EEZ99729.1 GFDF01001119 JAV12965.1 GQ390366 ACU77882.2
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000037510 UP000075886 UP000075920 UP000075900 UP000076408 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000078540 UP000008237 UP000001819 UP000007798 UP000008744 UP000075880 UP000053097 UP000279307 UP000268350 UP000005205 UP000192221 UP000075885 UP000037069 UP000001070 UP000069272 UP000009192 UP000030765 UP000007755 UP000076502 UP000008792 UP000000803 UP000092443 UP000092460 UP000007801 UP000092444 UP000000311 UP000008711 UP000092445 UP000078200 UP000002282 UP000245037 UP000000304 UP000095301 UP000000673 UP000036403 UP000007266
UP000037510 UP000075886 UP000075920 UP000075900 UP000076408 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000078540 UP000008237 UP000001819 UP000007798 UP000008744 UP000075880 UP000053097 UP000279307 UP000268350 UP000005205 UP000192221 UP000075885 UP000037069 UP000001070 UP000069272 UP000009192 UP000030765 UP000007755 UP000076502 UP000008792 UP000000803 UP000092443 UP000092460 UP000007801 UP000092444 UP000000311 UP000008711 UP000092445 UP000078200 UP000002282 UP000245037 UP000000304 UP000095301 UP000000673 UP000036403 UP000007266
Interpro
IPR005962
Tyr_3_mOase
+ More
IPR036329 Aro-AA_hydroxylase_C_sf
IPR019773 Tyrosine_3-monooxygenase-like
IPR001273 ArAA_hydroxylase
IPR019774 Aromatic-AA_hydroxylase_C
IPR036951 ArAA_hydroxylase_sf
IPR018301 ArAA_hydroxylase_Fe/CU_BS
IPR002912 ACT_dom
IPR041903 Eu_TyrOH_cat
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR020849 Small_GTPase_Ras-type
IPR036329 Aro-AA_hydroxylase_C_sf
IPR019773 Tyrosine_3-monooxygenase-like
IPR001273 ArAA_hydroxylase
IPR019774 Aromatic-AA_hydroxylase_C
IPR036951 ArAA_hydroxylase_sf
IPR018301 ArAA_hydroxylase_Fe/CU_BS
IPR002912 ACT_dom
IPR041903 Eu_TyrOH_cat
IPR005225 Small_GTP-bd_dom
IPR001806 Small_GTPase
IPR027417 P-loop_NTPase
IPR020849 Small_GTPase_Ras-type
Gene 3D
CDD
ProteinModelPortal
B7XAK2
E9L3G5
E9L3H1
E9L3J0
E9L3H7
E9L3J1
+ More
E9L3J2 E9L3G7 E9L3G4 E9L3H0 E9L3H5 E9L3H6 E9L3H8 E9L3I0 E7CZD7 A0A2H1VJX9 A5YVV1 A0A2W1BU82 M4GVG2 A0A2A4K6A9 A0A0G3VI81 Q402L8 D5MRL1 Q0E9S3 A0A194PT63 D5MRK5 D5LPT9 A6BLY5 F2VLL9 A0A212FDY4 D9IX57 A0A0L7L763 T1TCT2 A0A0G3VFY8 Q0E9S2 A0A182QXT4 A0A182WKJ2 A0A182RPF1 A0A182YQC3 A0A182V967 A0A182KPE6 A0A182X777 Q7PII7 A0A182I1Q6 A0A195BCD3 E2BF32 T1DPG9 Q29DR7 B4N2X5 B4H7A4 A0A182IZD0 A0A026WHN9 A0A3B0KGP5 A0A158NFE6 A0A1W4UXG4 E9IJ93 A0A182PJJ2 A0A0L0CQQ4 B4J096 A0A182FDH1 B4KW72 A0A084W0Q1 F4WGX8 A0A1Q3FU03 A5YVV2 A0A154P317 B4LHH8 B5X0J4 P18459 A0A1A9XWL5 F8J2H7 A0A1B0B587 B3M9H5 A0A336LN07 A0A1B0FBP0 E2A6B6 B3NFM6 A0A1A9Z3A0 A0A1A9VYP6 A0A0J9RQ65 B4PJR3 A0A2P8YCA1 B4QJT1 A0A0C9RB22 T1PBG7 G8E099 A0A1Y1M337 W5JBY1 A0A0Q9X3P9 W8BHQ1 A0A0A9WVH7 A0A034VJ95 A0A2R8G1S9 A0A0A1XT79 A0A0J7L2G0 A0A1W4VAQ6 A0A0K8VQZ7 D6WHA4 A0A1L8E390 C8CE48
E9L3J2 E9L3G7 E9L3G4 E9L3H0 E9L3H5 E9L3H6 E9L3H8 E9L3I0 E7CZD7 A0A2H1VJX9 A5YVV1 A0A2W1BU82 M4GVG2 A0A2A4K6A9 A0A0G3VI81 Q402L8 D5MRL1 Q0E9S3 A0A194PT63 D5MRK5 D5LPT9 A6BLY5 F2VLL9 A0A212FDY4 D9IX57 A0A0L7L763 T1TCT2 A0A0G3VFY8 Q0E9S2 A0A182QXT4 A0A182WKJ2 A0A182RPF1 A0A182YQC3 A0A182V967 A0A182KPE6 A0A182X777 Q7PII7 A0A182I1Q6 A0A195BCD3 E2BF32 T1DPG9 Q29DR7 B4N2X5 B4H7A4 A0A182IZD0 A0A026WHN9 A0A3B0KGP5 A0A158NFE6 A0A1W4UXG4 E9IJ93 A0A182PJJ2 A0A0L0CQQ4 B4J096 A0A182FDH1 B4KW72 A0A084W0Q1 F4WGX8 A0A1Q3FU03 A5YVV2 A0A154P317 B4LHH8 B5X0J4 P18459 A0A1A9XWL5 F8J2H7 A0A1B0B587 B3M9H5 A0A336LN07 A0A1B0FBP0 E2A6B6 B3NFM6 A0A1A9Z3A0 A0A1A9VYP6 A0A0J9RQ65 B4PJR3 A0A2P8YCA1 B4QJT1 A0A0C9RB22 T1PBG7 G8E099 A0A1Y1M337 W5JBY1 A0A0Q9X3P9 W8BHQ1 A0A0A9WVH7 A0A034VJ95 A0A2R8G1S9 A0A0A1XT79 A0A0J7L2G0 A0A1W4VAQ6 A0A0K8VQZ7 D6WHA4 A0A1L8E390 C8CE48
PDB
1TOH
E-value=1.12025e-122,
Score=1127
Ontologies
PATHWAY
GO
GO:0009072
GO:0042423
GO:0005506
GO:0004511
GO:0007165
GO:0003924
GO:0005525
GO:0016020
GO:0048085
GO:0048082
GO:0008049
GO:0035220
GO:0009611
GO:2000274
GO:0043052
GO:0040040
GO:0008344
GO:0042542
GO:0042416
GO:0042417
GO:0048066
GO:0006584
GO:0048067
GO:0042136
GO:0007619
GO:0030424
GO:0043204
GO:0005737
GO:0006585
GO:0016714
GO:0004497
GO:0006810
GO:0007169
GO:0016021
GO:0055114
PANTHER
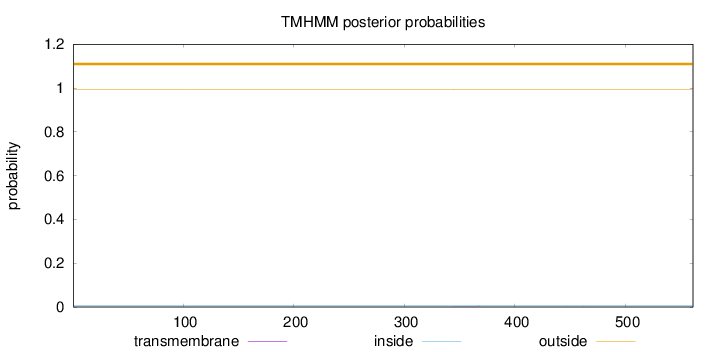
Topology
Length:
561
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01204
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00625
outside
1 - 561
Population Genetic Test Statistics
Pi
0.651827
Theta
5.993226
Tajima's D
-2.530351
CLR
18.693244
CSRT
0.000149992500374981
Interpretation
Possibly Positive selection
