Pre Gene Modal
BGIBMGA000564
Annotation
PREDICTED:_endoplasmic_reticulum_mannosyl-oligosaccharide_1?2-alpha-mannosidase_[Amyelois_transitella]
Full name
alpha-1,2-Mannosidase
Location in the cell
Mitochondrial Reliability : 1.384
Sequence
CDS
ATGAGACAGCAAAACTTAGAGACACATCTAAATTTTAATATAAATAATCATACAAATAATGTTAATGATAGAAAATGGGTCACCAATGGAGAGACAGCAATATTACTTAATGATCCAGCAATATCACATACTAAAATTATAAGGAAATGGAATCGTCTATCACGTCTTCAGAGATCCATAGTGTATGCTGTGCTAGTTGTGACTGCAACAGCTCTTTTGTTGCTGTATTTTTCTAAAACTGGTATTGTCATCATTGAGCGAGGCAATGAAGATTCAGTTCAGGAAAAATTAAGAAACACATCTCTAAATGCCAAACCTGCATTGGACAACACAGTTCAAATAAAATATGATCCTACAGTGTATGAGAGGCCTGTTTTTACAGGACCAAAAAATGATCGGCAACGGGCATTGGTGGAAAGTTTTAGACATGCATGGAGAGGGTATAAAGAGCATGCTTGGGGCCATGACAATCTAAAACCAGTCTCTGGTATGGCTTCTGACTGGTTCTCTCTTGGACTTACAATTGTTGACAGTTTGGATACAGCTTACATAATGGGACTAACTGATGTATTTGAGGAAGGCAAGCAATGGATCAAAGAAGAACTAATATTCACAAAAAATAAAGATGTTAATTTCTTTGAAGTCACCATTAGAATACTAGGTGGCTTGTTAACAAATCATTATTTCACTCAAGACCAGCTATTTCTTGATAAAGCTATTGACCTCGGAGACCGTCTCATGGCAGCATTTTCGTCACCATCTGGCATCCCATACTCTGATGTGAACTTGGGAGCTAGAACTGCTCATGCTCCTGAATGGTCACACTACAGCACCACAGCTGAAGTTACTACTGTGCAGTTGGAGTTTAGGCAACTGTCCAGAGCTAGTGGCAACCCTATATTCGAGGAGGCAGCAGCAGCCGTTTCCGAGAAGATTCATCAGCTACCAAAAAAACATGGACTTGTTCCAATATTTATTAATCCGAATACAGGACATTTTCTGCCTCACGCCACCATCACTCTCGGAGCAAGAGGGGATAGTTATTATGAATACCTGCTAAAACAATGGCTCCAGACTGGGAAAACAATCAACTATTTACTGGATGATTATTTAACCGGCATAGAAGGTGTACGTGAATTTTTGGCAAAACGTTCATCACCTAATAAACGATTGTTCATCGGAGAGTTGTCTGCTGGTTCAGAGGCTTTCAATCCGAAAATGGATCATTTGACCTGTTTCTTGCCTGGAACATTGGCCTTAGGTCATTTGAATGGCCTCCCCGACTGGCACATGACAATGGCCGAGGAACTTCTCTACACTTGCTATTTAACTTATGCTGCACATCCAACGTTTTTAGCTCCAGAAATAACACATTTCAATCTCGCGAGCACAACAGATGATATGTATACGAAATCGGCCGATGCTCACAGCTTATTGCGACCGGAATTTATTGAGAGTTTATGGTATATGTATCAAATAACTGGCAATACTACCTATCAAGAATGGGGCTGGCAGATATTCCAGGGCTTCGAAAAATATGCCAAAGTGCCAAACGGCTATTCGTCTCTAACAAACGTGAAATCTGAAAATCCAATACCACGTGACATGATGGAATCTTTCTTTTTGTCAGAAACACTTAAGTACTTGTACTTATTATTGAGTGACGATCGCTTTCTCATCGATTTGAATAAGGAGGGAGAGCGAGCTAAAGTTATCACTGGATGTTAG
Protein
MRQQNLETHLNFNINNHTNNVNDRKWVTNGETAILLNDPAISHTKIIRKWNRLSRLQRSIVYAVLVVTATALLLLYFSKTGIVIIERGNEDSVQEKLRNTSLNAKPALDNTVQIKYDPTVYERPVFTGPKNDRQRALVESFRHAWRGYKEHAWGHDNLKPVSGMASDWFSLGLTIVDSLDTAYIMGLTDVFEEGKQWIKEELIFTKNKDVNFFEVTIRILGGLLTNHYFTQDQLFLDKAIDLGDRLMAAFSSPSGIPYSDVNLGARTAHAPEWSHYSTTAEVTTVQLEFRQLSRASGNPIFEEAAAAVSEKIHQLPKKHGLVPIFINPNTGHFLPHATITLGARGDSYYEYLLKQWLQTGKTINYLLDDYLTGIEGVREFLAKRSSPNKRLFIGELSAGSEAFNPKMDHLTCFLPGTLALGHLNGLPDWHMTMAEELLYTCYLTYAAHPTFLAPEITHFNLASTTDDMYTKSADAHSLLRPEFIESLWYMYQITGNTTYQEWGWQIFQGFEKYAKVPNGYSSLTNVKSENPIPRDMMESFFLSETLKYLYLLLSDDRFLIDLNKEGERAKVITGC
Summary
Similarity
Belongs to the glycosyl hydrolase 47 family.
Feature
chain alpha-1,2-Mannosidase
Uniprot
H9ITI4
A0A2H1VLI6
A0A2A4K4X4
A0A194PRT3
A0A212FDT0
S4PDV8
+ More
A0A194QUM3 D6WGJ6 A0A2J7PHK4 A0A067QYZ1 A0A2P8YCE3 A0A1Y1LYG8 A0A1W4X3L7 A0A232FD64 K7IW08 A0A0L7R8Q0 N6T0C4 U4UAU2 A0A0N0BKY8 E0VZD5 V9IM31 E2BM44 A0A087ZXK6 E9G8U4 A0A154P1F5 A0A151IAG3 E2A3G3 A0A195EWI5 A0A195DRV4 F4WV42 A0A026W225 A0A195BTT8 A0A158NNQ2 W5JSD2 A0A2M4BHA7 A0A336M3E3 A0A0J7KUS3 A0A0P4VUB2 A0A0V0G8Z4 A0A336M3F3 A0A182FQ99 A0A023F409 A0A0A9XFA4 A0A182MVJ5 A0A2M4BN05 A0A2M3Z1W2 A0A0P5C3U2 A0A2M3ZG63 A0A1Q3F198 A0A1Q3F1G6 A0A182J968 A0A1B6E7L9 A0A069DZB3 A0A1Q3F1D7 T1IFB6 A0A182SEL3 A0A0P5F668 A0A0P5LVX3 A0A224XM47 A0A0P5KN18 A0A182VQB7 A0A2M4AL54 A0A084WQX2 A0A0P6D9Z0 A0A0P5R2K6 B0W9Q6 A0A182PIY8 A0A182XX80 A0A182RBU6 A0A182HCF3 A0A182WYF5 A0A182VG18 A0A182I8J8 A0A182LAF7 F5HIW5 A0A182GJ00 A0A182TE55 Q7QK05 A0A182N6X4 A0A182QZL9 A0A182KFC0 A0A0P5K4Q7 Q177N4 A0A1S4FCD0 U5ETE3 A0A1J1HVW9 A0A151WUR4 A0A1L8DR46 A0A1L8DQV9 A0A1B6L084 A0A1I8PBE7 I2C0B6 A0A1J1HU32 A0A1I8MB47 A0A1I8MB53 B4M5P2
A0A194QUM3 D6WGJ6 A0A2J7PHK4 A0A067QYZ1 A0A2P8YCE3 A0A1Y1LYG8 A0A1W4X3L7 A0A232FD64 K7IW08 A0A0L7R8Q0 N6T0C4 U4UAU2 A0A0N0BKY8 E0VZD5 V9IM31 E2BM44 A0A087ZXK6 E9G8U4 A0A154P1F5 A0A151IAG3 E2A3G3 A0A195EWI5 A0A195DRV4 F4WV42 A0A026W225 A0A195BTT8 A0A158NNQ2 W5JSD2 A0A2M4BHA7 A0A336M3E3 A0A0J7KUS3 A0A0P4VUB2 A0A0V0G8Z4 A0A336M3F3 A0A182FQ99 A0A023F409 A0A0A9XFA4 A0A182MVJ5 A0A2M4BN05 A0A2M3Z1W2 A0A0P5C3U2 A0A2M3ZG63 A0A1Q3F198 A0A1Q3F1G6 A0A182J968 A0A1B6E7L9 A0A069DZB3 A0A1Q3F1D7 T1IFB6 A0A182SEL3 A0A0P5F668 A0A0P5LVX3 A0A224XM47 A0A0P5KN18 A0A182VQB7 A0A2M4AL54 A0A084WQX2 A0A0P6D9Z0 A0A0P5R2K6 B0W9Q6 A0A182PIY8 A0A182XX80 A0A182RBU6 A0A182HCF3 A0A182WYF5 A0A182VG18 A0A182I8J8 A0A182LAF7 F5HIW5 A0A182GJ00 A0A182TE55 Q7QK05 A0A182N6X4 A0A182QZL9 A0A182KFC0 A0A0P5K4Q7 Q177N4 A0A1S4FCD0 U5ETE3 A0A1J1HVW9 A0A151WUR4 A0A1L8DR46 A0A1L8DQV9 A0A1B6L084 A0A1I8PBE7 I2C0B6 A0A1J1HU32 A0A1I8MB47 A0A1I8MB53 B4M5P2
EC Number
3.2.1.-
Pubmed
19121390
26354079
22118469
23622113
18362917
19820115
+ More
24845553 29403074 28004739 28648823 20075255 23537049 20566863 20798317 21292972 21719571 24508170 30249741 21347285 20920257 23761445 27129103 25474469 25401762 26823975 26334808 24438588 25244985 26483478 20966253 12364791 14747013 17210077 17510324 22561073 25315136 17994087
24845553 29403074 28004739 28648823 20075255 23537049 20566863 20798317 21292972 21719571 24508170 30249741 21347285 20920257 23761445 27129103 25474469 25401762 26823975 26334808 24438588 25244985 26483478 20966253 12364791 14747013 17210077 17510324 22561073 25315136 17994087
EMBL
BABH01006924
ODYU01003221
SOQ41685.1
NWSH01000125
PCG79305.1
KQ459594
+ More
KPI96156.1 AGBW02008997 OWR51916.1 GAIX01004792 JAA87768.1 KQ461108 KPJ09228.1 KQ971321 EFA00169.1 NEVH01025136 PNF15809.1 KK853182 KDR10175.1 PYGN01000706 PSN41913.1 GEZM01043996 JAV78592.1 NNAY01000450 OXU28307.1 KQ414631 KOC67257.1 APGK01049519 KB741156 ENN73509.1 KB632169 ERL89428.1 KQ435689 KOX81268.1 DS235851 EEB18741.1 JR052778 AEY61782.1 GL449153 EFN83230.1 GL732535 EFX84035.1 KQ434796 KZC05687.1 KQ978260 KYM95874.1 GL436428 EFN72027.1 KQ981953 KYN32511.1 KQ980530 KYN15650.1 GL888384 EGI61930.1 KK107485 QOIP01000006 EZA50112.1 RLU21984.1 KQ976417 KYM89846.1 ADTU01021714 ADTU01021715 ADMH02000490 ETN66213.1 GGFJ01003299 MBW52440.1 UFQT01000496 SSX24764.1 LBMM01002905 KMQ94202.1 GDKW01000333 JAI56262.1 GECL01001548 JAP04576.1 SSX24765.1 GBBI01003008 JAC15704.1 GBHO01024985 GDHC01016361 JAG18619.1 JAQ02268.1 AXCM01005046 GGFJ01005180 MBW54321.1 GGFM01001677 MBW22428.1 GDIP01180127 JAJ43275.1 GGFM01006771 MBW27522.1 GFDL01013715 JAV21330.1 GFDL01013642 JAV21403.1 GEDC01003356 JAS33942.1 GBGD01000940 JAC87949.1 GFDL01013691 JAV21354.1 ACPB03000966 GDIP01151237 LRGB01002451 JAJ72165.1 KZS07663.1 GDIQ01173461 JAK78264.1 GFTR01007325 JAW09101.1 GDIQ01181952 JAK69773.1 GGFK01008199 MBW41520.1 ATLV01025791 KE525400 KFB52616.1 GDIQ01253833 GDIQ01251147 GDIQ01244134 GDIQ01224645 GDIQ01214301 GDIQ01208157 GDIQ01205401 GDIQ01202443 GDIQ01171914 GDIQ01145825 GDIQ01117785 GDIQ01112434 GDIQ01101627 GDIQ01093484 GDIQ01082089 GDIQ01067800 JAN12648.1 GDIQ01106600 JAL45126.1 DS231865 EDS40375.1 JXUM01127240 KQ567165 KXJ69584.1 APCN01003043 AAAB01008805 EGK96226.1 JXUM01012498 KQ560355 KXJ82872.1 EAA03885.4 AXCN02000311 GDIQ01189600 JAK62125.1 CH477373 EAT42384.1 GANO01002820 JAB57051.1 CVRI01000021 CRK91508.1 KQ982745 KYQ51385.1 GFDF01005274 JAV08810.1 GFDF01005275 JAV08809.1 GEBQ01022922 JAT17055.1 JQ804932 AFJ59949.1 CRK91509.1 CH940652 EDW58968.1
KPI96156.1 AGBW02008997 OWR51916.1 GAIX01004792 JAA87768.1 KQ461108 KPJ09228.1 KQ971321 EFA00169.1 NEVH01025136 PNF15809.1 KK853182 KDR10175.1 PYGN01000706 PSN41913.1 GEZM01043996 JAV78592.1 NNAY01000450 OXU28307.1 KQ414631 KOC67257.1 APGK01049519 KB741156 ENN73509.1 KB632169 ERL89428.1 KQ435689 KOX81268.1 DS235851 EEB18741.1 JR052778 AEY61782.1 GL449153 EFN83230.1 GL732535 EFX84035.1 KQ434796 KZC05687.1 KQ978260 KYM95874.1 GL436428 EFN72027.1 KQ981953 KYN32511.1 KQ980530 KYN15650.1 GL888384 EGI61930.1 KK107485 QOIP01000006 EZA50112.1 RLU21984.1 KQ976417 KYM89846.1 ADTU01021714 ADTU01021715 ADMH02000490 ETN66213.1 GGFJ01003299 MBW52440.1 UFQT01000496 SSX24764.1 LBMM01002905 KMQ94202.1 GDKW01000333 JAI56262.1 GECL01001548 JAP04576.1 SSX24765.1 GBBI01003008 JAC15704.1 GBHO01024985 GDHC01016361 JAG18619.1 JAQ02268.1 AXCM01005046 GGFJ01005180 MBW54321.1 GGFM01001677 MBW22428.1 GDIP01180127 JAJ43275.1 GGFM01006771 MBW27522.1 GFDL01013715 JAV21330.1 GFDL01013642 JAV21403.1 GEDC01003356 JAS33942.1 GBGD01000940 JAC87949.1 GFDL01013691 JAV21354.1 ACPB03000966 GDIP01151237 LRGB01002451 JAJ72165.1 KZS07663.1 GDIQ01173461 JAK78264.1 GFTR01007325 JAW09101.1 GDIQ01181952 JAK69773.1 GGFK01008199 MBW41520.1 ATLV01025791 KE525400 KFB52616.1 GDIQ01253833 GDIQ01251147 GDIQ01244134 GDIQ01224645 GDIQ01214301 GDIQ01208157 GDIQ01205401 GDIQ01202443 GDIQ01171914 GDIQ01145825 GDIQ01117785 GDIQ01112434 GDIQ01101627 GDIQ01093484 GDIQ01082089 GDIQ01067800 JAN12648.1 GDIQ01106600 JAL45126.1 DS231865 EDS40375.1 JXUM01127240 KQ567165 KXJ69584.1 APCN01003043 AAAB01008805 EGK96226.1 JXUM01012498 KQ560355 KXJ82872.1 EAA03885.4 AXCN02000311 GDIQ01189600 JAK62125.1 CH477373 EAT42384.1 GANO01002820 JAB57051.1 CVRI01000021 CRK91508.1 KQ982745 KYQ51385.1 GFDF01005274 JAV08810.1 GFDF01005275 JAV08809.1 GEBQ01022922 JAT17055.1 JQ804932 AFJ59949.1 CRK91509.1 CH940652 EDW58968.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000007266
+ More
UP000235965 UP000027135 UP000245037 UP000192223 UP000215335 UP000002358 UP000053825 UP000019118 UP000030742 UP000053105 UP000009046 UP000008237 UP000005203 UP000000305 UP000076502 UP000078542 UP000000311 UP000078541 UP000078492 UP000007755 UP000053097 UP000279307 UP000078540 UP000005205 UP000000673 UP000036403 UP000069272 UP000075883 UP000075880 UP000015103 UP000075901 UP000076858 UP000075920 UP000030765 UP000002320 UP000075885 UP000076408 UP000075900 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000075882 UP000007062 UP000075902 UP000075884 UP000075886 UP000075881 UP000008820 UP000183832 UP000075809 UP000095300 UP000095301 UP000008792
UP000235965 UP000027135 UP000245037 UP000192223 UP000215335 UP000002358 UP000053825 UP000019118 UP000030742 UP000053105 UP000009046 UP000008237 UP000005203 UP000000305 UP000076502 UP000078542 UP000000311 UP000078541 UP000078492 UP000007755 UP000053097 UP000279307 UP000078540 UP000005205 UP000000673 UP000036403 UP000069272 UP000075883 UP000075880 UP000015103 UP000075901 UP000076858 UP000075920 UP000030765 UP000002320 UP000075885 UP000076408 UP000075900 UP000069940 UP000249989 UP000076407 UP000075903 UP000075840 UP000075882 UP000007062 UP000075902 UP000075884 UP000075886 UP000075881 UP000008820 UP000183832 UP000075809 UP000095300 UP000095301 UP000008792
PRIDE
Pfam
PF01532 Glyco_hydro_47
Interpro
SUPFAM
SSF48225
SSF48225
Gene 3D
ProteinModelPortal
H9ITI4
A0A2H1VLI6
A0A2A4K4X4
A0A194PRT3
A0A212FDT0
S4PDV8
+ More
A0A194QUM3 D6WGJ6 A0A2J7PHK4 A0A067QYZ1 A0A2P8YCE3 A0A1Y1LYG8 A0A1W4X3L7 A0A232FD64 K7IW08 A0A0L7R8Q0 N6T0C4 U4UAU2 A0A0N0BKY8 E0VZD5 V9IM31 E2BM44 A0A087ZXK6 E9G8U4 A0A154P1F5 A0A151IAG3 E2A3G3 A0A195EWI5 A0A195DRV4 F4WV42 A0A026W225 A0A195BTT8 A0A158NNQ2 W5JSD2 A0A2M4BHA7 A0A336M3E3 A0A0J7KUS3 A0A0P4VUB2 A0A0V0G8Z4 A0A336M3F3 A0A182FQ99 A0A023F409 A0A0A9XFA4 A0A182MVJ5 A0A2M4BN05 A0A2M3Z1W2 A0A0P5C3U2 A0A2M3ZG63 A0A1Q3F198 A0A1Q3F1G6 A0A182J968 A0A1B6E7L9 A0A069DZB3 A0A1Q3F1D7 T1IFB6 A0A182SEL3 A0A0P5F668 A0A0P5LVX3 A0A224XM47 A0A0P5KN18 A0A182VQB7 A0A2M4AL54 A0A084WQX2 A0A0P6D9Z0 A0A0P5R2K6 B0W9Q6 A0A182PIY8 A0A182XX80 A0A182RBU6 A0A182HCF3 A0A182WYF5 A0A182VG18 A0A182I8J8 A0A182LAF7 F5HIW5 A0A182GJ00 A0A182TE55 Q7QK05 A0A182N6X4 A0A182QZL9 A0A182KFC0 A0A0P5K4Q7 Q177N4 A0A1S4FCD0 U5ETE3 A0A1J1HVW9 A0A151WUR4 A0A1L8DR46 A0A1L8DQV9 A0A1B6L084 A0A1I8PBE7 I2C0B6 A0A1J1HU32 A0A1I8MB47 A0A1I8MB53 B4M5P2
A0A194QUM3 D6WGJ6 A0A2J7PHK4 A0A067QYZ1 A0A2P8YCE3 A0A1Y1LYG8 A0A1W4X3L7 A0A232FD64 K7IW08 A0A0L7R8Q0 N6T0C4 U4UAU2 A0A0N0BKY8 E0VZD5 V9IM31 E2BM44 A0A087ZXK6 E9G8U4 A0A154P1F5 A0A151IAG3 E2A3G3 A0A195EWI5 A0A195DRV4 F4WV42 A0A026W225 A0A195BTT8 A0A158NNQ2 W5JSD2 A0A2M4BHA7 A0A336M3E3 A0A0J7KUS3 A0A0P4VUB2 A0A0V0G8Z4 A0A336M3F3 A0A182FQ99 A0A023F409 A0A0A9XFA4 A0A182MVJ5 A0A2M4BN05 A0A2M3Z1W2 A0A0P5C3U2 A0A2M3ZG63 A0A1Q3F198 A0A1Q3F1G6 A0A182J968 A0A1B6E7L9 A0A069DZB3 A0A1Q3F1D7 T1IFB6 A0A182SEL3 A0A0P5F668 A0A0P5LVX3 A0A224XM47 A0A0P5KN18 A0A182VQB7 A0A2M4AL54 A0A084WQX2 A0A0P6D9Z0 A0A0P5R2K6 B0W9Q6 A0A182PIY8 A0A182XX80 A0A182RBU6 A0A182HCF3 A0A182WYF5 A0A182VG18 A0A182I8J8 A0A182LAF7 F5HIW5 A0A182GJ00 A0A182TE55 Q7QK05 A0A182N6X4 A0A182QZL9 A0A182KFC0 A0A0P5K4Q7 Q177N4 A0A1S4FCD0 U5ETE3 A0A1J1HVW9 A0A151WUR4 A0A1L8DR46 A0A1L8DQV9 A0A1B6L084 A0A1I8PBE7 I2C0B6 A0A1J1HU32 A0A1I8MB47 A0A1I8MB53 B4M5P2
PDB
1FMI
E-value=1.3037e-133,
Score=1222
Ontologies
PATHWAY
GO
Topology
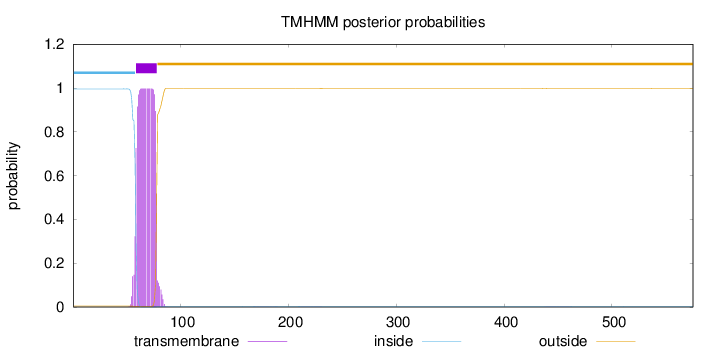
Length:
575
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.16918
Exp number, first 60 AAs:
2.31505
Total prob of N-in:
0.99593
inside
1 - 58
TMhelix
59 - 78
outside
79 - 575
Population Genetic Test Statistics
Pi
0.796795
Theta
4.145084
Tajima's D
-2.168107
CLR
0.009041
CSRT
0.00534973251337433
Interpretation
Uncertain
