Gene
KWMTBOMO00239 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000649
Annotation
ATP_synthase_b_[Operophtera_brumata]
Full name
ATP synthase subunit b, mitochondrial
+ More
Gustatory receptor
Gustatory receptor
Alternative Name
FO-ATP synthase subunit b
Location in the cell
Cytoplasmic Reliability : 1.69 Mitochondrial Reliability : 2.053
Sequence
CDS
ATGATTTTTAGGCCTTTTATTCAACCGAGGTTCAATAAACAGTTTCTATTCGTGCCCAATTTTATACTAGCTCATACCAGCACTTGTCCAGCTTCAAAACCCAAATGTGGAGCATCCGCAGGCAAGGGGGGAACGACTTCTATAAAAAAAATTGATTTCAAACGCGCTGAAAAATCTGCCCCATGTCGATTCGGTTTCATACCTGAAGAATGGTTTCAATTTTTTCACTCAAAAACGGGCGTTACTGGACCATATATATTTGGACTAGTTTTTGTTAATTATCTTGTCAGCAAAGAAATATATGTGTTAGAACATGAATATTACAGTGGGCTATCGTATGTTGTTATTTTGTACTTTATATCGAAAAAAATGGCCCCTGGAATAGGAGCTAGCCTTGACAAAGAAGTAGACGCCGAAATAGCCGAGTGGGAGAAGGGGAGAGCAGATCAAATGAAGGTTTTTGAAACCACGATTAAAGATGCTAAAGATGCGCAATGGAGAGCTGAAGGCCAAAAAATATTGATCGAAGCCAAGAAAGAGAACGTGGCTATGCAATTAGAAGCAGTATATCGAGAAAGAGCAATGCGGTTATACCAAATGGTTAAAGGGAGATTAGATTATCACGTCAAAAAACGGAGAACTGAAAACAAATTACACCAGAAATGGATGATTGCATGGATTTTGGAGAATGTTCATAAATCAATAACAGCGGATTTTCAGAAACAAGCCCTTAATCAGGCGATTAAAGATTTAGCTCTTGCCGCTAGTCGTGTTAAATAG
Protein
MIFRPFIQPRFNKQFLFVPNFILAHTSTCPASKPKCGASAGKGGTTSIKKIDFKRAEKSAPCRFGFIPEEWFQFFHSKTGVTGPYIFGLVFVNYLVSKEIYVLEHEYYSGLSYVVILYFISKKMAPGIGASLDKEVDAEIAEWEKGRADQMKVFETTIKDAKDAQWRAEGQKILIEAKKENVAMQLEAVYRERAMRLYQMVKGRLDYHVKKRRTENKLHQKWMIAWILENVHKSITADFQKQALNQAIKDLALAASRVK
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the eukaryotic ATPase B chain family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Keywords
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transport
Feature
chain ATP synthase subunit b, mitochondrial
Uniprot
A0A0L7LU08
A0A1E1VYD1
A0A194R036
A0A2A4JM46
A0A2W1BKJ7
S4NX19
+ More
A0A2H1WRD0 Q8ITF0 A0A2A4JR94 A0A194RE02 I4DNJ3 A0A2H1WII3 H9JGM3 A0A182UYT9 A0A182XUM4 Q7PWZ7 A0A182IID8 A0A1E1W304 Q2F5T4 A0A1Z3GD36 B0WFJ7 A0A182UAX8 I4DMM3 A0A220K8M5 A0A3G1T1F7 A0A182QL16 A0A194PTU2 A0A212FKR5 A0A2W1BW64 A0A0K8T0I9 B8YDH6 A0A1Q3FJZ6 T1E9T8 A0A182PR71 A0A182FRZ6 A0A2M4BYC5 A0A182YNB9 A0A2M3Z0Q4 A0A1Y9H2V7 A0A2M4A5I2 A0A2M4A2G4 A0A2M4A4I6 W5J8P6 A0A182RG08 A0A2M3ZG00 A0A084W069 A0A182T4Y1 A0A182JCW2 A0A2M4CT31 A0A1J1HZV6 T1DIJ6 A0A182WR21 T1PAZ0 A0A1L8E0X9 A0A1I8PCL5 U5EXN0 A0A0L0CKY8 Q179J9 A0A1L8EF93 A0A023EKM8 W8BTA8 A0A0K8W8C5 A0A034WG46 A0A0C9RGA1 B3MA73 B4J2P5 A0A0K8WIH7 A0A1W4UKX2 A0A3F2YUX3 B4HLA2 B4QNN6 Q94516 Q2LZ91 B4HAF5 A0A182JRG7 B3NGL2 A0A182MJN5 A0A3B0JZF9 B4PEC8 A0A1B0AID5 D3TR96 B4LGA6 A0A182GHQ5 B4N4A0 A0A1A9YNJ7 A0A1A9WG23 A0A1A9VAR8 A0A0M4EZP6 A0A1B0CZQ2 A0A1B0B956 A0A2J7R1Y3 V5H551 B4L0K7 R4WR37 J3JWD6 W8S2I1 A0A3L8DVU7 A0A232FGN5 E2ADM9
A0A2H1WRD0 Q8ITF0 A0A2A4JR94 A0A194RE02 I4DNJ3 A0A2H1WII3 H9JGM3 A0A182UYT9 A0A182XUM4 Q7PWZ7 A0A182IID8 A0A1E1W304 Q2F5T4 A0A1Z3GD36 B0WFJ7 A0A182UAX8 I4DMM3 A0A220K8M5 A0A3G1T1F7 A0A182QL16 A0A194PTU2 A0A212FKR5 A0A2W1BW64 A0A0K8T0I9 B8YDH6 A0A1Q3FJZ6 T1E9T8 A0A182PR71 A0A182FRZ6 A0A2M4BYC5 A0A182YNB9 A0A2M3Z0Q4 A0A1Y9H2V7 A0A2M4A5I2 A0A2M4A2G4 A0A2M4A4I6 W5J8P6 A0A182RG08 A0A2M3ZG00 A0A084W069 A0A182T4Y1 A0A182JCW2 A0A2M4CT31 A0A1J1HZV6 T1DIJ6 A0A182WR21 T1PAZ0 A0A1L8E0X9 A0A1I8PCL5 U5EXN0 A0A0L0CKY8 Q179J9 A0A1L8EF93 A0A023EKM8 W8BTA8 A0A0K8W8C5 A0A034WG46 A0A0C9RGA1 B3MA73 B4J2P5 A0A0K8WIH7 A0A1W4UKX2 A0A3F2YUX3 B4HLA2 B4QNN6 Q94516 Q2LZ91 B4HAF5 A0A182JRG7 B3NGL2 A0A182MJN5 A0A3B0JZF9 B4PEC8 A0A1B0AID5 D3TR96 B4LGA6 A0A182GHQ5 B4N4A0 A0A1A9YNJ7 A0A1A9WG23 A0A1A9VAR8 A0A0M4EZP6 A0A1B0CZQ2 A0A1B0B956 A0A2J7R1Y3 V5H551 B4L0K7 R4WR37 J3JWD6 W8S2I1 A0A3L8DVU7 A0A232FGN5 E2ADM9
Pubmed
26227816
26354079
28756777
23622113
22651552
19121390
+ More
12364791 14747013 17210077 28630352 22118469 25244985 20920257 23761445 24438588 24330624 25315136 26108605 17510324 24945155 24495485 25348373 17994087 20966253 22936249 10731132 12537572 10071211 15632085 17550304 20353571 26483478 23691247 22516182 30249741 28648823 20798317
12364791 14747013 17210077 28630352 22118469 25244985 20920257 23761445 24438588 24330624 25315136 26108605 17510324 24945155 24495485 25348373 17994087 20966253 22936249 10731132 12537572 10071211 15632085 17550304 20353571 26483478 23691247 22516182 30249741 28648823 20798317
EMBL
JTDY01000090
KOB78937.1
GDQN01011301
JAT79753.1
KQ461108
KPJ09211.1
+ More
NWSH01001094 PCG72654.1 KZ150110 PZC73366.1 GAIX01012337 JAA80223.1 ODYU01010448 SOQ55567.1 AF518015 AAN39695.1 NWSH01000785 PCG74228.1 KQ460367 KPJ15510.1 AK402964 KQ459589 BAM19483.1 KPI97660.1 ODYU01008799 SOQ52712.1 BABH01034379 AAAB01008987 EAA01034.4 APCN01000816 GDQN01009745 JAT81309.1 DQ311339 ABD36283.1 KY783434 ASC55682.1 DS231918 EDS26265.1 AK402541 BAM19163.1 MF319726 ASJ26440.1 MG992364 AXY94802.1 AXCN02001485 KQ459594 KPI96174.1 AGBW02007996 OWR54337.1 KZ149929 PZC77447.1 GBRD01006900 JAG58921.1 FJ524854 ACL77780.1 GFDL01007239 JAV27806.1 GAMD01001022 JAB00569.1 GGFJ01008918 MBW58059.1 GGFM01001325 MBW22076.1 GGFK01002753 MBW36074.1 GGFK01001662 MBW34983.1 GGFK01002348 MBW35669.1 ADMH02002101 ETN59169.1 GGFM01006688 MBW27439.1 ATLV01019092 KE525262 KFB43613.1 GGFL01004334 MBW68512.1 CVRI01000037 CRK93499.1 GALA01000896 JAA93956.1 KA645926 AFP60555.1 GFDF01001723 JAV12361.1 GANO01000872 JAB58999.1 JRES01000254 KNC32945.1 CH477349 EAT42891.1 GFDG01001596 JAV17203.1 GAPW01003860 JAC09738.1 GAMC01001960 JAC04596.1 GDHF01018934 GDHF01005050 JAI33380.1 JAI47264.1 GAKP01004391 JAC54561.1 GBYB01006076 JAG75843.1 CH902618 EDV39087.1 CH916366 EDV96036.1 GDHF01001460 JAI50854.1 CH480815 EDW40921.1 CM000363 CM002912 EDX09903.1 KMY98746.1 AE014296 BT004878 BT011453 Y08967 CH379069 EAL29617.1 CH479240 EDW37553.1 CH954178 EDV51248.1 AXCM01000086 OUUW01000012 SPP87467.1 CM000159 EDW92966.1 CCAG010023354 EZ423948 ADD20224.1 CH940647 EDW69414.1 JXUM01064333 KQ562293 KXJ76220.1 CH964095 EDW78974.1 CP012525 ALC44225.1 AJVK01020937 JXJN01010282 NEVH01008202 PNF34847.1 GALX01000543 JAB67923.1 CH933809 EDW18084.1 AK417122 BAN20337.1 BT127554 AEE62516.1 KJ001158 AHL84105.1 QOIP01000003 RLU24442.1 NNAY01000229 OXU29855.1 GL438820 EFN68391.1
NWSH01001094 PCG72654.1 KZ150110 PZC73366.1 GAIX01012337 JAA80223.1 ODYU01010448 SOQ55567.1 AF518015 AAN39695.1 NWSH01000785 PCG74228.1 KQ460367 KPJ15510.1 AK402964 KQ459589 BAM19483.1 KPI97660.1 ODYU01008799 SOQ52712.1 BABH01034379 AAAB01008987 EAA01034.4 APCN01000816 GDQN01009745 JAT81309.1 DQ311339 ABD36283.1 KY783434 ASC55682.1 DS231918 EDS26265.1 AK402541 BAM19163.1 MF319726 ASJ26440.1 MG992364 AXY94802.1 AXCN02001485 KQ459594 KPI96174.1 AGBW02007996 OWR54337.1 KZ149929 PZC77447.1 GBRD01006900 JAG58921.1 FJ524854 ACL77780.1 GFDL01007239 JAV27806.1 GAMD01001022 JAB00569.1 GGFJ01008918 MBW58059.1 GGFM01001325 MBW22076.1 GGFK01002753 MBW36074.1 GGFK01001662 MBW34983.1 GGFK01002348 MBW35669.1 ADMH02002101 ETN59169.1 GGFM01006688 MBW27439.1 ATLV01019092 KE525262 KFB43613.1 GGFL01004334 MBW68512.1 CVRI01000037 CRK93499.1 GALA01000896 JAA93956.1 KA645926 AFP60555.1 GFDF01001723 JAV12361.1 GANO01000872 JAB58999.1 JRES01000254 KNC32945.1 CH477349 EAT42891.1 GFDG01001596 JAV17203.1 GAPW01003860 JAC09738.1 GAMC01001960 JAC04596.1 GDHF01018934 GDHF01005050 JAI33380.1 JAI47264.1 GAKP01004391 JAC54561.1 GBYB01006076 JAG75843.1 CH902618 EDV39087.1 CH916366 EDV96036.1 GDHF01001460 JAI50854.1 CH480815 EDW40921.1 CM000363 CM002912 EDX09903.1 KMY98746.1 AE014296 BT004878 BT011453 Y08967 CH379069 EAL29617.1 CH479240 EDW37553.1 CH954178 EDV51248.1 AXCM01000086 OUUW01000012 SPP87467.1 CM000159 EDW92966.1 CCAG010023354 EZ423948 ADD20224.1 CH940647 EDW69414.1 JXUM01064333 KQ562293 KXJ76220.1 CH964095 EDW78974.1 CP012525 ALC44225.1 AJVK01020937 JXJN01010282 NEVH01008202 PNF34847.1 GALX01000543 JAB67923.1 CH933809 EDW18084.1 AK417122 BAN20337.1 BT127554 AEE62516.1 KJ001158 AHL84105.1 QOIP01000003 RLU24442.1 NNAY01000229 OXU29855.1 GL438820 EFN68391.1
Proteomes
UP000037510
UP000053240
UP000218220
UP000053268
UP000005204
UP000075903
+ More
UP000076407 UP000007062 UP000075840 UP000002320 UP000075902 UP000075886 UP000007151 UP000075885 UP000069272 UP000076408 UP000075884 UP000000673 UP000075900 UP000030765 UP000075901 UP000075880 UP000183832 UP000075920 UP000095301 UP000095300 UP000037069 UP000008820 UP000007801 UP000001070 UP000192221 UP000075882 UP000001292 UP000000304 UP000000803 UP000001819 UP000008744 UP000075881 UP000008711 UP000075883 UP000268350 UP000002282 UP000092445 UP000092444 UP000008792 UP000069940 UP000249989 UP000007798 UP000092443 UP000091820 UP000078200 UP000092553 UP000092462 UP000092460 UP000235965 UP000009192 UP000279307 UP000215335 UP000000311
UP000076407 UP000007062 UP000075840 UP000002320 UP000075902 UP000075886 UP000007151 UP000075885 UP000069272 UP000076408 UP000075884 UP000000673 UP000075900 UP000030765 UP000075901 UP000075880 UP000183832 UP000075920 UP000095301 UP000095300 UP000037069 UP000008820 UP000007801 UP000001070 UP000192221 UP000075882 UP000001292 UP000000304 UP000000803 UP000001819 UP000008744 UP000075881 UP000008711 UP000075883 UP000268350 UP000002282 UP000092445 UP000092444 UP000008792 UP000069940 UP000249989 UP000007798 UP000092443 UP000091820 UP000078200 UP000092553 UP000092462 UP000092460 UP000235965 UP000009192 UP000279307 UP000215335 UP000000311
Interpro
Gene 3D
ProteinModelPortal
A0A0L7LU08
A0A1E1VYD1
A0A194R036
A0A2A4JM46
A0A2W1BKJ7
S4NX19
+ More
A0A2H1WRD0 Q8ITF0 A0A2A4JR94 A0A194RE02 I4DNJ3 A0A2H1WII3 H9JGM3 A0A182UYT9 A0A182XUM4 Q7PWZ7 A0A182IID8 A0A1E1W304 Q2F5T4 A0A1Z3GD36 B0WFJ7 A0A182UAX8 I4DMM3 A0A220K8M5 A0A3G1T1F7 A0A182QL16 A0A194PTU2 A0A212FKR5 A0A2W1BW64 A0A0K8T0I9 B8YDH6 A0A1Q3FJZ6 T1E9T8 A0A182PR71 A0A182FRZ6 A0A2M4BYC5 A0A182YNB9 A0A2M3Z0Q4 A0A1Y9H2V7 A0A2M4A5I2 A0A2M4A2G4 A0A2M4A4I6 W5J8P6 A0A182RG08 A0A2M3ZG00 A0A084W069 A0A182T4Y1 A0A182JCW2 A0A2M4CT31 A0A1J1HZV6 T1DIJ6 A0A182WR21 T1PAZ0 A0A1L8E0X9 A0A1I8PCL5 U5EXN0 A0A0L0CKY8 Q179J9 A0A1L8EF93 A0A023EKM8 W8BTA8 A0A0K8W8C5 A0A034WG46 A0A0C9RGA1 B3MA73 B4J2P5 A0A0K8WIH7 A0A1W4UKX2 A0A3F2YUX3 B4HLA2 B4QNN6 Q94516 Q2LZ91 B4HAF5 A0A182JRG7 B3NGL2 A0A182MJN5 A0A3B0JZF9 B4PEC8 A0A1B0AID5 D3TR96 B4LGA6 A0A182GHQ5 B4N4A0 A0A1A9YNJ7 A0A1A9WG23 A0A1A9VAR8 A0A0M4EZP6 A0A1B0CZQ2 A0A1B0B956 A0A2J7R1Y3 V5H551 B4L0K7 R4WR37 J3JWD6 W8S2I1 A0A3L8DVU7 A0A232FGN5 E2ADM9
A0A2H1WRD0 Q8ITF0 A0A2A4JR94 A0A194RE02 I4DNJ3 A0A2H1WII3 H9JGM3 A0A182UYT9 A0A182XUM4 Q7PWZ7 A0A182IID8 A0A1E1W304 Q2F5T4 A0A1Z3GD36 B0WFJ7 A0A182UAX8 I4DMM3 A0A220K8M5 A0A3G1T1F7 A0A182QL16 A0A194PTU2 A0A212FKR5 A0A2W1BW64 A0A0K8T0I9 B8YDH6 A0A1Q3FJZ6 T1E9T8 A0A182PR71 A0A182FRZ6 A0A2M4BYC5 A0A182YNB9 A0A2M3Z0Q4 A0A1Y9H2V7 A0A2M4A5I2 A0A2M4A2G4 A0A2M4A4I6 W5J8P6 A0A182RG08 A0A2M3ZG00 A0A084W069 A0A182T4Y1 A0A182JCW2 A0A2M4CT31 A0A1J1HZV6 T1DIJ6 A0A182WR21 T1PAZ0 A0A1L8E0X9 A0A1I8PCL5 U5EXN0 A0A0L0CKY8 Q179J9 A0A1L8EF93 A0A023EKM8 W8BTA8 A0A0K8W8C5 A0A034WG46 A0A0C9RGA1 B3MA73 B4J2P5 A0A0K8WIH7 A0A1W4UKX2 A0A3F2YUX3 B4HLA2 B4QNN6 Q94516 Q2LZ91 B4HAF5 A0A182JRG7 B3NGL2 A0A182MJN5 A0A3B0JZF9 B4PEC8 A0A1B0AID5 D3TR96 B4LGA6 A0A182GHQ5 B4N4A0 A0A1A9YNJ7 A0A1A9WG23 A0A1A9VAR8 A0A0M4EZP6 A0A1B0CZQ2 A0A1B0B956 A0A2J7R1Y3 V5H551 B4L0K7 R4WR37 J3JWD6 W8S2I1 A0A3L8DVU7 A0A232FGN5 E2ADM9
PDB
2CLY
E-value=7.21193e-35,
Score=366
Ontologies
PATHWAY
GO
PANTHER
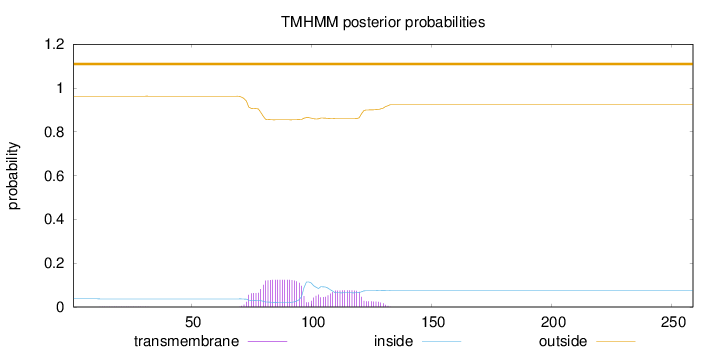
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Cell membrane
Mitochondrion inner membrane
Cell membrane
Length:
259
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
4.15752
Exp number, first 60 AAs:
0.00735
Total prob of N-in:
0.03722
outside
1 - 259
Population Genetic Test Statistics
Pi
0.908104
Theta
3.004967
Tajima's D
-1.780853
CLR
0
CSRT
0.0215989200539973
Interpretation
Possibly Positive selection
