Gene
KWMTBOMO00238 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000648
Annotation
imaginal_disk_growth_factor_[Bombyx_mori]
Full name
Chitinase-like protein EN03
+ More
Chitinase-like protein Idgf4
Chitinase-like protein Idgf4
Alternative Name
Imaginal disk growth factor-like protein
Imaginal disk growth factor protein 4
Imaginal disk growth factor protein 4
Location in the cell
Mitochondrial Reliability : 0.954
Sequence
CDS
ATGAAGCTATTTATCGCTCTAGTCGGGCTCTTGGCCCTCGCTAAGGCCCTACCTGCGGTAACCCACAGCAAAGTACTTTGTTACTACGACAGCAGGAGCTATGTCAGAGAATCTCAAGCCCGCATGCTGCCGTTGGACCTCGATCCCGCTCTGTCGTTCTGCACCCACTTGCTGTACGGCTATGCCGGTATCCAGCCTGACACCTATAAGCTGGTGTCCTTAAACGAGAACCTGGACATAGACCGAACACACGACAACTACCGTGCGATCACCAGCTTGAAAGCCAAGTACCCTGGTCTCACTGTATTATTATCTGTTGGTGGCGACGCTGATACCGAAGAACCAGAAAAATATAACCTTCTGCTGGAATCGCAGCAAGCCCGTACTGCTTTCATTAATTCCGGAGTGCTGTTGGCTGAACAATATGGTTTCGATGGAATTGACCTCGCCTGGCAGTTCCCAAGAGTTAAGCCTAAGAAGATCCGCTCGACCTGGGGATCGCTTTGGCATGGAATTAAGAAGACATTCGGCACCACGCCAGTCGATGAGAAGGAATCTGAGCACCGTGAAGGTTTCACTGCCCTTGTTCGTGAATTGAAACAGGCCCTTATCCACAAGCCTAAGATGCAGCTCGGCGTGACTGTTTTGCCCAACGTTAATTCTACAATTTACCACGACGTGCCCGCCATCATTAACTTAGTTGACTATGTAAACGTTGGCGCTTACGATTATTACACTCCGACACGCAACAACAAAGAAGCTGATTACACAGCTCCTATTTACACCCCACAAAATCGCAACCCACTGCAGAACGCCGACGCCGCTGTCACATACTGGCTCACGAGCGGTGCTCCAAGCCAAAAGATAGTATTGTCTATCGCGACCTTCGGTCGTACCTGGAAACTGGACGCTGACAGCGAAATTGCGGGAGTCCCTCCAATCCACACTGATGGTCCCGGCGAAGCTGGACCATACGTTAAGACTGAAGGTTTGCTCAGTTACCCAGAAGTCTGCGGTAAGTTGATCAACCCCAACCAGCAGAAGGGAATGCGTCCTCACCTCAGGAAGGTTACTGACCCCAGCAAGCGTTTCGGAACATACGCCTTCCGTCTTCCTGATGACAACGGTGAGGGTGGCATTTGGGTATCATACGAGGATCCTGATACTGCTGGCCAGAAAGCTGCTTACGTCAAGTCAAAGAATCTCGGTGGTGTCGCTATTGTGGACCTATCATTGGATGACTTCCGCGGTCTCTGTACCGGAGACAAGTATCCAATCCTTAGGGCCGCTAAATACCGTCTCTAA
Protein
MKLFIALVGLLALAKALPAVTHSKVLCYYDSRSYVRESQARMLPLDLDPALSFCTHLLYGYAGIQPDTYKLVSLNENLDIDRTHDNYRAITSLKAKYPGLTVLLSVGGDADTEEPEKYNLLLESQQARTAFINSGVLLAEQYGFDGIDLAWQFPRVKPKKIRSTWGSLWHGIKKTFGTTPVDEKESEHREGFTALVRELKQALIHKPKMQLGVTVLPNVNSTIYHDVPAIINLVDYVNVGAYDYYTPTRNNKEADYTAPIYTPQNRNPLQNADAAVTYWLTSGAPSQKIVLSIATFGRTWKLDADSEIAGVPPIHTDGPGEAGPYVKTEGLLSYPEVCGKLINPNQQKGMRPHLRKVTDPSKRFGTYAFRLPDDNGEGGIWVSYEDPDTAGQKAAYVKSKNLGGVAIVDLSLDDFRGLCTGDKYPILRAAKYRL
Summary
Description
Cooperates with insulin-like peptides to stimulate the proliferation, polarization and motility of imaginal disk cells. May act by stabilizing the binding of insulin-like peptides to its receptor through a simultaneous interaction with both molecules to form a multiprotein signaling complex (By similarity).
Cooperates with insulin-like peptides to stimulate the proliferation, polarization and motility of imaginal disk cells. May act by stabilizing the binding of insulin-like peptides to its receptor through a simultaneous interaction with both molecules to form a multiprotein signaling complex.
Cooperates with insulin-like peptides to stimulate the proliferation, polarization and motility of imaginal disk cells. May act by stabilizing the binding of insulin-like peptides to its receptor through a simultaneous interaction with both molecules to form a multiprotein signaling complex.
Miscellaneous
Lacks the typical Glu active site in position 152 that is replaced by a Gln residue, preventing the hydrolase activity. Its precise function remains unclear.
Lacks the typical Glu active site in position 158 that is replaced by a Gln residue, preventing the hydrolase activity. Its precise function remains unclear.
Lacks the typical Glu active site in position 156 that is replaced by a Gln residue, preventing the hydrolase activity. Its precise function remains unclear.
Lacks the typical Glu active site in position 158 that is replaced by a Gln residue, preventing the hydrolase activity. Its precise function remains unclear.
Lacks the typical Glu active site in position 156 that is replaced by a Gln residue, preventing the hydrolase activity. Its precise function remains unclear.
Similarity
Belongs to the glycosyl hydrolase 18 family.
Belongs to the glycosyl hydrolase 18 family. IDGF subfamily.
Belongs to the glycosyl hydrolase 18 family. IDGF subfamily.
Keywords
Complete proteome
Direct protein sequencing
Disulfide bond
Glycoprotein
Reference proteome
Secreted
Signal
Developmental protein
Feature
chain Chitinase-like protein EN03
Uniprot
A7BEX9
Q9GV28
H9ITR8
A0A2A4J8G1
D0EJV6
Q0QJJ5
+ More
A0A212FDU6 Q6IWP3 A0A2H1V6B4 T1WFZ2 S4NT73 A0A194QB02 I4DM57 A0A2W1BNW3 A0A194QZW8 A0A3G1T182 I4DJ57 A0JCK4 A0A1W4X8W6 U5EUT6 A0A1W4WZG9 A0A182TSA9 A0A182UUX7 A0NF62 A0A182JLL9 A0A182L1N9 Q6JEK4 A0A2C9GR73 A0A182QFT6 A0A182LRE7 A0A1B0EZB9 A0A182T6F3 A0A1A9WS71 V5I8T0 A0A2C9GR77 Q7Q3V3 A0A182G3Y5 Q6JEK5 A0A182VLG6 A0A023ESA4 A0A182TM46 A0A182L1N7 A0A0L0C2Z8 A0A2M3Z1L3 A0A182WU84 A0A1S4F0F9 A0A023ES96 A0A2M3ZEW9 A0A023ES78 A0A182P4X1 A0A023EUI5 A0A182JP37 A0A1L8E1S0 A0A1A9UEB2 A0A182Y035 A0A084VNB3 A0A182J7A7 A0A2I6Q778 A0A3B0J863 A0A1B0BCD4 T1DSM7 A0A1L8E1W9 Q17JL4 D3TLB4 Q2PQM7 A0A182NU84 A0A2P0P9I2 A0A1A9YS97 A0A2Z2Z1A0 A0A182RC98 B4JIS9 Q29FW8 A0A034W3G0 W8B3T8 A0A1B0ADI8 A0A182MPR3 A0A0K8V176 A0A0A1WM01 B4L2Y9 T1PF14 A0A2M4BQD2 B4M255 A0A182W789 B0W7I9 W5JAF2 X2JEB6 Q9W303 Q0Z935 A0A0M5J2Z6 Q0Z936 B4PXD8 A0A182G3Y4 A0A1W4VHQ8 A0A084VNB4 A0A2M4AAV2 T1EAM6 A0A2M4BMS1 A0A1Q3FJM1 A0A1B0FNT6 B3NT34
A0A212FDU6 Q6IWP3 A0A2H1V6B4 T1WFZ2 S4NT73 A0A194QB02 I4DM57 A0A2W1BNW3 A0A194QZW8 A0A3G1T182 I4DJ57 A0JCK4 A0A1W4X8W6 U5EUT6 A0A1W4WZG9 A0A182TSA9 A0A182UUX7 A0NF62 A0A182JLL9 A0A182L1N9 Q6JEK4 A0A2C9GR73 A0A182QFT6 A0A182LRE7 A0A1B0EZB9 A0A182T6F3 A0A1A9WS71 V5I8T0 A0A2C9GR77 Q7Q3V3 A0A182G3Y5 Q6JEK5 A0A182VLG6 A0A023ESA4 A0A182TM46 A0A182L1N7 A0A0L0C2Z8 A0A2M3Z1L3 A0A182WU84 A0A1S4F0F9 A0A023ES96 A0A2M3ZEW9 A0A023ES78 A0A182P4X1 A0A023EUI5 A0A182JP37 A0A1L8E1S0 A0A1A9UEB2 A0A182Y035 A0A084VNB3 A0A182J7A7 A0A2I6Q778 A0A3B0J863 A0A1B0BCD4 T1DSM7 A0A1L8E1W9 Q17JL4 D3TLB4 Q2PQM7 A0A182NU84 A0A2P0P9I2 A0A1A9YS97 A0A2Z2Z1A0 A0A182RC98 B4JIS9 Q29FW8 A0A034W3G0 W8B3T8 A0A1B0ADI8 A0A182MPR3 A0A0K8V176 A0A0A1WM01 B4L2Y9 T1PF14 A0A2M4BQD2 B4M255 A0A182W789 B0W7I9 W5JAF2 X2JEB6 Q9W303 Q0Z935 A0A0M5J2Z6 Q0Z936 B4PXD8 A0A182G3Y4 A0A1W4VHQ8 A0A084VNB4 A0A2M4AAV2 T1EAM6 A0A2M4BMS1 A0A1Q3FJM1 A0A1B0FNT6 B3NT34
Pubmed
11222941
11280994
19121390
7949257
16835019
22118469
+ More
15288202 23622113 26354079 22651552 28756777 30578597 12364791 14747013 17210077 20966253 15271211 26483478 24945155 26108605 25244985 24438588 29236932 17510324 20353571 16907828 17994087 15632085 25348373 24495485 25830018 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9847235 12537569 18342250 18362917 19820115 17550304
15288202 23622113 26354079 22651552 28756777 30578597 12364791 14747013 17210077 20966253 15271211 26483478 24945155 26108605 25244985 24438588 29236932 17510324 20353571 16907828 17994087 15632085 25348373 24495485 25830018 25315136 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 9847235 12537569 18342250 18362917 19820115 17550304
EMBL
AB183872
BAF73623.1
AB041634
BAB16695.1
BABH01006915
BABH01006916
+ More
NWSH01002459 PCG68251.1 GQ843826 ACW82749.1 DQ355162 ABC79625.1 AGBW02008997 OWR51911.1 AY616136 AAT36640.1 ODYU01000916 SOQ36395.1 KF040475 AGU16453.1 GAIX01012296 JAA80264.1 KQ459463 KPJ00601.1 AK402375 BAM18997.1 KZ150110 PZC73363.1 KQ461108 KPJ09136.1 MG846880 AXY94732.1 AK401325 BAM17947.1 MH899224 AB282642 AZS52300.1 BAF36822.1 GANO01002177 JAB57694.1 AAAB01008964 EAU76358.1 AY496421 AAS80138.1 APCN01002479 AXCN02000772 AXCM01008455 AJVK01007194 GALX01004150 JAB64316.1 EAA12233.3 JXUM01041493 KQ561285 KXJ79101.1 AY496420 AAS80137.1 GAPW01001475 JAC12123.1 JRES01001070 KNC25789.1 GGFM01001671 MBW22422.1 GAPW01001473 JAC12125.1 GGFM01006229 MBW26980.1 GAPW01001498 JAC12100.1 GAPW01001474 JAC12124.1 GFDF01001437 JAV12647.1 ATLV01014736 KE524984 KFB39457.1 KY274439 AUM84824.1 OUUW01000003 SPP78025.1 JXJN01011956 GAMD01001409 JAB00182.1 GFDF01001438 JAV12646.1 CH477232 EAT46826.1 EZ422216 ADD18492.1 DQ307196 KU739373 ARA90550.1 KY681046 AUP42573.1 CH916370 EDW00526.1 CH379065 EAL31461.3 GAKP01010100 MH250169 JAC48852.1 QAV56502.1 GAMC01010745 JAB95810.1 GDHF01019989 JAI32325.1 GBXI01014213 JAD00079.1 CH933810 EDW07875.2 KA646478 AFP61107.1 GGFJ01005837 MBW54978.1 CH940651 EDW65759.2 DS231854 EDS38024.1 ADMH02002034 ETN59840.1 AE014298 AHN59538.1 AF102239 AY070943 BT003236 DQ659254 KQ971342 ABG47452.1 EFA03804.1 CP012528 ALC48324.1 DQ659253 ABG47451.1 EFA03805.1 CM000162 EDX02889.1 JXUM01041491 JXUM01041492 KXJ79100.1 KFB39458.1 GGFK01004584 MBW37905.1 GAMD01000908 JAB00683.1 GGFJ01005181 MBW54322.1 GFDL01007276 JAV27769.1 CCAG010019910 CH954180 EDV46214.1
NWSH01002459 PCG68251.1 GQ843826 ACW82749.1 DQ355162 ABC79625.1 AGBW02008997 OWR51911.1 AY616136 AAT36640.1 ODYU01000916 SOQ36395.1 KF040475 AGU16453.1 GAIX01012296 JAA80264.1 KQ459463 KPJ00601.1 AK402375 BAM18997.1 KZ150110 PZC73363.1 KQ461108 KPJ09136.1 MG846880 AXY94732.1 AK401325 BAM17947.1 MH899224 AB282642 AZS52300.1 BAF36822.1 GANO01002177 JAB57694.1 AAAB01008964 EAU76358.1 AY496421 AAS80138.1 APCN01002479 AXCN02000772 AXCM01008455 AJVK01007194 GALX01004150 JAB64316.1 EAA12233.3 JXUM01041493 KQ561285 KXJ79101.1 AY496420 AAS80137.1 GAPW01001475 JAC12123.1 JRES01001070 KNC25789.1 GGFM01001671 MBW22422.1 GAPW01001473 JAC12125.1 GGFM01006229 MBW26980.1 GAPW01001498 JAC12100.1 GAPW01001474 JAC12124.1 GFDF01001437 JAV12647.1 ATLV01014736 KE524984 KFB39457.1 KY274439 AUM84824.1 OUUW01000003 SPP78025.1 JXJN01011956 GAMD01001409 JAB00182.1 GFDF01001438 JAV12646.1 CH477232 EAT46826.1 EZ422216 ADD18492.1 DQ307196 KU739373 ARA90550.1 KY681046 AUP42573.1 CH916370 EDW00526.1 CH379065 EAL31461.3 GAKP01010100 MH250169 JAC48852.1 QAV56502.1 GAMC01010745 JAB95810.1 GDHF01019989 JAI32325.1 GBXI01014213 JAD00079.1 CH933810 EDW07875.2 KA646478 AFP61107.1 GGFJ01005837 MBW54978.1 CH940651 EDW65759.2 DS231854 EDS38024.1 ADMH02002034 ETN59840.1 AE014298 AHN59538.1 AF102239 AY070943 BT003236 DQ659254 KQ971342 ABG47452.1 EFA03804.1 CP012528 ALC48324.1 DQ659253 ABG47451.1 EFA03805.1 CM000162 EDX02889.1 JXUM01041491 JXUM01041492 KXJ79100.1 KFB39458.1 GGFK01004584 MBW37905.1 GAMD01000908 JAB00683.1 GGFJ01005181 MBW54322.1 GFDL01007276 JAV27769.1 CCAG010019910 CH954180 EDV46214.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000192223
+ More
UP000075902 UP000075903 UP000007062 UP000075880 UP000075882 UP000075840 UP000075886 UP000075883 UP000092462 UP000075901 UP000091820 UP000069940 UP000249989 UP000037069 UP000076407 UP000075885 UP000075881 UP000078200 UP000076408 UP000030765 UP000268350 UP000092460 UP000008820 UP000092444 UP000075884 UP000092443 UP000075900 UP000001070 UP000001819 UP000092445 UP000009192 UP000095301 UP000008792 UP000075920 UP000002320 UP000000673 UP000000803 UP000007266 UP000092553 UP000002282 UP000192221 UP000008711
UP000075902 UP000075903 UP000007062 UP000075880 UP000075882 UP000075840 UP000075886 UP000075883 UP000092462 UP000075901 UP000091820 UP000069940 UP000249989 UP000037069 UP000076407 UP000075885 UP000075881 UP000078200 UP000076408 UP000030765 UP000268350 UP000092460 UP000008820 UP000092444 UP000075884 UP000092443 UP000075900 UP000001070 UP000001819 UP000092445 UP000009192 UP000095301 UP000008792 UP000075920 UP000002320 UP000000673 UP000000803 UP000007266 UP000092553 UP000002282 UP000192221 UP000008711
Pfam
PF00704 Glyco_hydro_18
Interpro
Gene 3D
ProteinModelPortal
A7BEX9
Q9GV28
H9ITR8
A0A2A4J8G1
D0EJV6
Q0QJJ5
+ More
A0A212FDU6 Q6IWP3 A0A2H1V6B4 T1WFZ2 S4NT73 A0A194QB02 I4DM57 A0A2W1BNW3 A0A194QZW8 A0A3G1T182 I4DJ57 A0JCK4 A0A1W4X8W6 U5EUT6 A0A1W4WZG9 A0A182TSA9 A0A182UUX7 A0NF62 A0A182JLL9 A0A182L1N9 Q6JEK4 A0A2C9GR73 A0A182QFT6 A0A182LRE7 A0A1B0EZB9 A0A182T6F3 A0A1A9WS71 V5I8T0 A0A2C9GR77 Q7Q3V3 A0A182G3Y5 Q6JEK5 A0A182VLG6 A0A023ESA4 A0A182TM46 A0A182L1N7 A0A0L0C2Z8 A0A2M3Z1L3 A0A182WU84 A0A1S4F0F9 A0A023ES96 A0A2M3ZEW9 A0A023ES78 A0A182P4X1 A0A023EUI5 A0A182JP37 A0A1L8E1S0 A0A1A9UEB2 A0A182Y035 A0A084VNB3 A0A182J7A7 A0A2I6Q778 A0A3B0J863 A0A1B0BCD4 T1DSM7 A0A1L8E1W9 Q17JL4 D3TLB4 Q2PQM7 A0A182NU84 A0A2P0P9I2 A0A1A9YS97 A0A2Z2Z1A0 A0A182RC98 B4JIS9 Q29FW8 A0A034W3G0 W8B3T8 A0A1B0ADI8 A0A182MPR3 A0A0K8V176 A0A0A1WM01 B4L2Y9 T1PF14 A0A2M4BQD2 B4M255 A0A182W789 B0W7I9 W5JAF2 X2JEB6 Q9W303 Q0Z935 A0A0M5J2Z6 Q0Z936 B4PXD8 A0A182G3Y4 A0A1W4VHQ8 A0A084VNB4 A0A2M4AAV2 T1EAM6 A0A2M4BMS1 A0A1Q3FJM1 A0A1B0FNT6 B3NT34
A0A212FDU6 Q6IWP3 A0A2H1V6B4 T1WFZ2 S4NT73 A0A194QB02 I4DM57 A0A2W1BNW3 A0A194QZW8 A0A3G1T182 I4DJ57 A0JCK4 A0A1W4X8W6 U5EUT6 A0A1W4WZG9 A0A182TSA9 A0A182UUX7 A0NF62 A0A182JLL9 A0A182L1N9 Q6JEK4 A0A2C9GR73 A0A182QFT6 A0A182LRE7 A0A1B0EZB9 A0A182T6F3 A0A1A9WS71 V5I8T0 A0A2C9GR77 Q7Q3V3 A0A182G3Y5 Q6JEK5 A0A182VLG6 A0A023ESA4 A0A182TM46 A0A182L1N7 A0A0L0C2Z8 A0A2M3Z1L3 A0A182WU84 A0A1S4F0F9 A0A023ES96 A0A2M3ZEW9 A0A023ES78 A0A182P4X1 A0A023EUI5 A0A182JP37 A0A1L8E1S0 A0A1A9UEB2 A0A182Y035 A0A084VNB3 A0A182J7A7 A0A2I6Q778 A0A3B0J863 A0A1B0BCD4 T1DSM7 A0A1L8E1W9 Q17JL4 D3TLB4 Q2PQM7 A0A182NU84 A0A2P0P9I2 A0A1A9YS97 A0A2Z2Z1A0 A0A182RC98 B4JIS9 Q29FW8 A0A034W3G0 W8B3T8 A0A1B0ADI8 A0A182MPR3 A0A0K8V176 A0A0A1WM01 B4L2Y9 T1PF14 A0A2M4BQD2 B4M255 A0A182W789 B0W7I9 W5JAF2 X2JEB6 Q9W303 Q0Z935 A0A0M5J2Z6 Q0Z936 B4PXD8 A0A182G3Y4 A0A1W4VHQ8 A0A084VNB4 A0A2M4AAV2 T1EAM6 A0A2M4BMS1 A0A1Q3FJM1 A0A1B0FNT6 B3NT34
PDB
1JNE
E-value=1.68266e-115,
Score=1064
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 16,
Likelihood: 0.998295
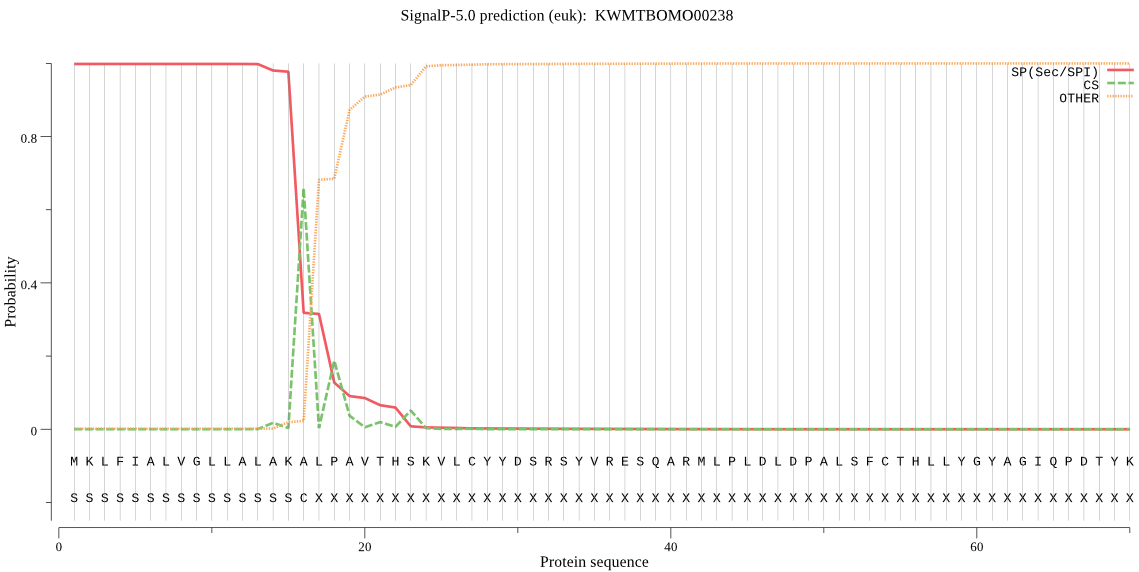
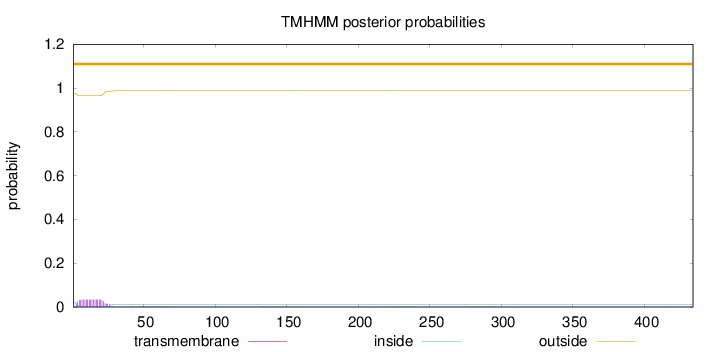
Length:
434
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.745189999999999
Exp number, first 60 AAs:
0.7265
Total prob of N-in:
0.02397
outside
1 - 434
Population Genetic Test Statistics
Pi
0.714414
Theta
4.758097
Tajima's D
-2.452972
CLR
0.014224
CSRT
0.000299985000749962
Interpretation
Possibly Positive selection
