Gene
KWMTBOMO00234
Pre Gene Modal
BGIBMGA000645
Annotation
PREDICTED:_glutamate_receptor_1-like_[Bombyx_mori]
Full name
Glutamate receptor 1
Alternative Name
Glutamate receptor I
Kainate-selective glutamate receptor
Kainate-selective glutamate receptor
Location in the cell
PlasmaMembrane Reliability : 3.565
Sequence
CDS
ATGCTAACTATAAATATTCGGCGTTTGATTGTTCGTGAACATGTGCTGCATACATGTAGCATTGTCTGCAATCAATTCGGACGTGGGGTAATGGCGATGCTGGGTGCTGTAACTCCGGACTCCTTCGACACTCTGCACTCGTATTCCAACACCTTCCAGATGCCGTTCGTGACCCCCTGGTTTCCTGAAAAGGTTATACCACCGTCATCGGGCCTCATCGATTATGCCGTCAGCATGCGACCGGACTACCACCGAGCTGTCATCGACACCATCACCTTCTATGGTTGGAAGAACGTCATATACGTGTATGATTCCCACGACGGTCTGCTTCGCCTCCAGCAACTGTACCAAAGTCTGCAACCGGGCAACGCTACTTTTCGAATTTCTAATGTGAAACGCGTCTCCAACGCAACTGACGTGATCGCATTTCTCGGCTCGATCGAGCGTCTGGATCGCTGGAGCAATAAATACGTTGTACTCGACTCCACAACACAACTCGCTAAAGAAGCTCTAATTTTGCACGTGCGCGACGTAAGGCTCGGGCGCCGGAACTACCATTATTTTCTAAGCGGTCTGGTGATGGACGATCGCTGGGAAAGGGAGGTCACAGAATTCGGAGCTATTAACATAACTGGGTTTAGAGTACTGGATTTTTCGAGAAAAGTCGTAAGAGATTTTATCGATGTTTGGAAGAAGAATTCTATTTCGGCCCAAGCTGCCCTCATGTACGACGCGGTCCAGGTGTTGGTGGACGCCATAGTGCGGCTACTCCGTAAGAAGCCAGATATACTGAGGGGAACGATAAGACGGAATGCCAACGTGAACACAAGCCGCATCATCGACTGCAACCCTAAAGGCAAAATTACACCTTACGAACACGGCGACAAGATATCACGGATGATTAAAAAGACGGAGTTAGATGGGCTGACCGGAAGCCTTCGGTTCAACGAAGAGGGCCATCGTCGCAACTTTACCTTGCAAATCATGGAGCTCACTGTTGATGGTGAAATGGTCAAAATTGCGACGTGGTATGACAACCGCGGACTTGTGCCGGTGGTGCCCAAGCTATCACCATTGTCTCCCCCCGGAACATACGATCGAAACAAAACATACATAGTGTCCACCATAGAAGAGCCCCCATACGTTATGCGGCATAAAGCAGGCGGGCCGGAATCCACGCCTAACGACCCATACAGAGGCTTCTGTGTAGACCTCGCCAAAATGTTATCGGATAAGTTGGAAATCAAATATGACTTACGACTGGTGAAAGACGGAAAGTACGGAAACGACAATCCGAAAATAGTTGGCGGTTGGGACGGGATGGTTGGCGAACTCGTCAGGAAGGAAGTAGATATGGCTATAGCTCCACTCACGATCACACTAGAAAGAGAAACTGTTATTGACTTTAGCAAACCATTTTTGTCGTTCGATGTCAAAACGAAAAAAAATTCATTCAAGGACCCCACTACGATATTCAGTTTTCTTCATCCACTTTCTAAGGAAATATGGCTCTGCATTATATTCAGCTTGTTCGCCGTTAGTGTTGTACTATTTCTGGTATCGAGATTTTCCCCGCACGAATGGCGTTTGATTTCATTCACCGACTCTCAAACTTCGGATCACTCGGAAGTGGCAACAACAAAGACGACCGCGGTCAACGAATTCACCTTCTGGAATTCTATGTGGTTTTCGTTGGGTTCTTTTATGCAGCAGGGCAGCGACATTACGCCGAGATCGATTTCCGGAAGAATTGTAGGATCGGTTTGGTGGTTTTTCGCATTGATCGTTATCTCGTCGTACACTGCCAATTTGGCGTCTTACCTCACCCTGACACGTATATCGGAACCGAGTCAATCATACAACAAATTAGCGACATGCCCTGAGGATACGGCTGGCAATCTAGGAAATTCAATGTTCCAAGAAGGAGGTGAAGGGAATGGCTGGCTTAATTTTATCTTCGATCCTGCTGGTTCTAATAGCGAAAATGATAAAAAGCCCTGTGAAATGTTAGTCACCGTTGCCAGCACTGGGGTGCAGGATTTTGCAGTCGCAATGCCGAAGGGATCCAAATTGCGGGACGGAATCAACCTTGCCCTCCAATCACTTAAAAATGACGGCGAGTTGCAAAAACTCATCCGCAAATGGTTTACTAAAGCAGAGTGTGAAGCTAACCAGGACGTTCAAGGCTCGGAACTGACTTTAGGTCAAGTGGCGGGACTGTTCTACGTCTTGCTGGGTGGACTGGGTCTTGCTTTAGGCGTGGCATTATTGGAATTTTGTCAGCATGGTCGTGCTGAAGCAGCTCGCGCCAATGTCCCACTAGGGGCAGCGTTACGCGCCCGCGCTCAACTTGCTTCGTCTGGAACCAACCGTGACAGGAAGAACAATTCTCAGCGAACCCCACAACGTGATAGCGACCGACTCGGATGGAATGGCGGAGCCTTCACTGGAGTGAGCTACTACAGCCCGGCCACCCAGATCGGTCAGGAGGAGACTTCCCTGCACGCTAGCTTCACGCAAGTCTGA
Protein
MLTINIRRLIVREHVLHTCSIVCNQFGRGVMAMLGAVTPDSFDTLHSYSNTFQMPFVTPWFPEKVIPPSSGLIDYAVSMRPDYHRAVIDTITFYGWKNVIYVYDSHDGLLRLQQLYQSLQPGNATFRISNVKRVSNATDVIAFLGSIERLDRWSNKYVVLDSTTQLAKEALILHVRDVRLGRRNYHYFLSGLVMDDRWEREVTEFGAINITGFRVLDFSRKVVRDFIDVWKKNSISAQAALMYDAVQVLVDAIVRLLRKKPDILRGTIRRNANVNTSRIIDCNPKGKITPYEHGDKISRMIKKTELDGLTGSLRFNEEGHRRNFTLQIMELTVDGEMVKIATWYDNRGLVPVVPKLSPLSPPGTYDRNKTYIVSTIEEPPYVMRHKAGGPESTPNDPYRGFCVDLAKMLSDKLEIKYDLRLVKDGKYGNDNPKIVGGWDGMVGELVRKEVDMAIAPLTITLERETVIDFSKPFLSFDVKTKKNSFKDPTTIFSFLHPLSKEIWLCIIFSLFAVSVVLFLVSRFSPHEWRLISFTDSQTSDHSEVATTKTTAVNEFTFWNSMWFSLGSFMQQGSDITPRSISGRIVGSVWWFFALIVISSYTANLASYLTLTRISEPSQSYNKLATCPEDTAGNLGNSMFQEGGEGNGWLNFIFDPAGSNSENDKKPCEMLVTVASTGVQDFAVAMPKGSKLRDGINLALQSLKNDGELQKLIRKWFTKAECEANQDVQGSELTLGQVAGLFYVLLGGLGLALGVALLEFCQHGRAEAARANVPLGAALRARAQLASSGTNRDRKNNSQRTPQRDSDRLGWNGGAFTGVSYYSPATQIGQEETSLHASFTQV
Summary
Description
Receptor for glutamate. L-glutamate acts as an excitatory neurotransmitter at many synapses in the central nervous system. The postsynaptic actions of Glu are mediated by a variety of receptors that are named according to their selective agonists (By similarity). Forms ligand-gated ion channels which are activated by kainate.
Subunit
Homooligomer.
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Keywords
3D-structure
Alternative splicing
Cell junction
Cell membrane
Complete proteome
Glycoprotein
Ion channel
Ion transport
Ligand-gated ion channel
Membrane
Postsynaptic cell membrane
Receptor
Reference proteome
Signal
Synapse
Transmembrane
Transmembrane helix
Transport
Feature
chain Glutamate receptor 1
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9ITR5
A0A0V0J107
A0A2W1BAM5
A0A212EMP7
A0A2H1VKE5
A0A194QVZ8
+ More
A0A194Q6H5 A0A194QUJ7 A0A212EMS3 A0A0V0J2T4 A0A2Y9D1L1 D7EHZ7 A0A1S4G7Q1 A0A075T6Z5 A0A1Y1N186 Q7PNT0 A0A3L8DF15 A0A158NFE2 T1HDK9 A0A151JZ81 A0A0C9QXE8 A0A1J1J876 B4KVF4 B4LIB6 A0A1W4UXQ3 B4IXW4 B3M9H9 A0A0R3P8X0 B3NGJ4 A0A3B0KNZ8 A0A336LRY5 B4N2Y2 Q29DR4 B4PJQ8 B4QJS5 Q03445 A0A1L2YXN2 A0A0M4EXY0 E9H8R8 A0A0P6AXH5 A0A0P4ZYN3 A0A0P5BVK8 A0A0P5C8P6 A0A0P6IP12 A0A1L2YXM6 A0A0P5GK06 A0A0C9RSE8 A0A0P5EM22 A0A0P5BI92 A0A151XFV4 A0A0P5BKB1 A0A0P5CZ00 A0A2H8TG59 A0A194Q4Z3 A0A0P5ALF8 A0A0P4ZRV5 A0A0P6DQ48 A0A0P5HB37 A0A0N8A5P0 E0VW33 A0A0N8ADX7 A0A0P5CJN3 A0A0P6HD12 A0A0P5IK63 A0A0N8BR75 A0A0P4YWX8 A0A0P6DJM2 A0A182KPE3 A0A0P5JIQ7 A0A2S2PJV1 A0A182JJ78 A0A0P5BHC1 A0A0M8ZMS8 A0A0P5BKU0 Q170E0 A0A0L7R971 A0A154P0S2 E2BF27 A0A0N7ZQJ7 B4H799 A0A195BCD1 A0A087ZQD0 A0A195E8E3 A0A0K2T188 A0A0P5SK38 E2A6A6 A0A0P4XW61 A0A026WHJ2 T1JZC1 A0A0M4EDF2 A0A2A4ISK2 F4WUH2 A0A336MVV7
A0A194Q6H5 A0A194QUJ7 A0A212EMS3 A0A0V0J2T4 A0A2Y9D1L1 D7EHZ7 A0A1S4G7Q1 A0A075T6Z5 A0A1Y1N186 Q7PNT0 A0A3L8DF15 A0A158NFE2 T1HDK9 A0A151JZ81 A0A0C9QXE8 A0A1J1J876 B4KVF4 B4LIB6 A0A1W4UXQ3 B4IXW4 B3M9H9 A0A0R3P8X0 B3NGJ4 A0A3B0KNZ8 A0A336LRY5 B4N2Y2 Q29DR4 B4PJQ8 B4QJS5 Q03445 A0A1L2YXN2 A0A0M4EXY0 E9H8R8 A0A0P6AXH5 A0A0P4ZYN3 A0A0P5BVK8 A0A0P5C8P6 A0A0P6IP12 A0A1L2YXM6 A0A0P5GK06 A0A0C9RSE8 A0A0P5EM22 A0A0P5BI92 A0A151XFV4 A0A0P5BKB1 A0A0P5CZ00 A0A2H8TG59 A0A194Q4Z3 A0A0P5ALF8 A0A0P4ZRV5 A0A0P6DQ48 A0A0P5HB37 A0A0N8A5P0 E0VW33 A0A0N8ADX7 A0A0P5CJN3 A0A0P6HD12 A0A0P5IK63 A0A0N8BR75 A0A0P4YWX8 A0A0P6DJM2 A0A182KPE3 A0A0P5JIQ7 A0A2S2PJV1 A0A182JJ78 A0A0P5BHC1 A0A0M8ZMS8 A0A0P5BKU0 Q170E0 A0A0L7R971 A0A154P0S2 E2BF27 A0A0N7ZQJ7 B4H799 A0A195BCD1 A0A087ZQD0 A0A195E8E3 A0A0K2T188 A0A0P5SK38 E2A6A6 A0A0P4XW61 A0A026WHJ2 T1JZC1 A0A0M4EDF2 A0A2A4ISK2 F4WUH2 A0A336MVV7
Pubmed
EMBL
BABH01006903
BABH01006904
BABH01006905
GDKB01000027
JAP38469.1
KZ150215
+ More
PZC72138.1 AGBW02013810 OWR42780.1 ODYU01003020 SOQ41261.1 KQ461108 KPJ09140.1 KQ459463 KPJ00605.1 KPJ09142.1 OWR42778.1 GDKB01000026 JAP38470.1 DS497669 EFA12935.1 KF768730 AIG51924.1 GEZM01016690 JAV91188.1 AAAB01008960 EAA11672.5 QOIP01000009 RLU18508.1 ADTU01014325 ADTU01014326 ADTU01014327 ADTU01014328 ADTU01014329 ADTU01014330 ADTU01014331 ADTU01014332 ADTU01014333 ADTU01014334 ACPB03000938 ACPB03000939 ACPB03000940 ACPB03000941 ACPB03000942 ACPB03000943 ACPB03000944 ACPB03000945 ACPB03000946 ACPB03000947 KQ981410 KYN41902.1 GBYB01005332 JAG75099.1 CVRI01000075 CRL08584.1 CH933809 EDW18397.1 CH940647 EDW69683.2 CH916366 EDV96485.1 CH902618 EDV41192.1 CH379070 KRT08845.1 CH954178 EDV51160.1 OUUW01000012 SPP87596.1 UFQT01000066 SSX19329.1 CH964062 EDW78721.1 EAL30350.1 CM000159 EDW93657.1 CM000363 CM002912 EDX09466.1 KMY97953.1 M97192 AE014296 BT003218 BT050435 KX016777 APF30026.1 CP012525 ALC43237.1 GL732605 EFX71861.1 GDIP01029649 JAM74066.1 GDIP01209509 JAJ13893.1 GDIP01180016 JAJ43386.1 GDIP01174566 JAJ48836.1 GDIQ01008113 JAN86624.1 KX016772 APF30021.1 GDIQ01241334 JAK10391.1 GBYB01011575 JAG81342.1 GDIP01146122 JAJ77280.1 GDIP01184715 JAJ38687.1 KQ982182 KYQ59168.1 GDIP01183366 JAJ40036.1 GDIP01163482 JAJ59920.1 GFXV01000473 MBW12278.1 KPJ00608.1 GDIP01197266 JAJ26136.1 GDIP01209510 JAJ13892.1 GDIQ01073743 GDIQ01052945 JAN20994.1 GDIQ01230429 JAK21296.1 GDIP01180017 JAJ43385.1 DS235817 EEB17589.1 GDIP01156921 JAJ66481.1 GDIP01174565 JAJ48837.1 GDIQ01021126 JAN73611.1 GDIQ01211392 JAK40333.1 GDIQ01152859 JAK98866.1 GDIP01222360 JAJ01042.1 GDIQ01076542 JAN18195.1 GDIQ01202352 JAK49373.1 GGMR01017110 MBY29729.1 GDIP01184716 JAJ38686.1 KQ436160 KOX67415.1 GDIP01183367 JAJ40035.1 CH477476 EAT40294.1 KQ414627 KOC67420.1 KQ434795 KZC05526.1 GL447910 EFN85705.1 GDIP01222361 JAJ01041.1 CH479217 EDW33705.1 KQ976514 KYM82208.1 KQ979440 KYN21485.1 HACA01001900 CDW19261.1 GDIP01138773 JAL64941.1 GL437123 EFN70991.1 GDIP01235911 JAI87490.1 KK107199 EZA55532.1 CAEY01001120 ALC43621.1 NWSH01007937 PCG62745.1 GL888365 EGI62137.1 UFQT01002179 SSX32863.1
PZC72138.1 AGBW02013810 OWR42780.1 ODYU01003020 SOQ41261.1 KQ461108 KPJ09140.1 KQ459463 KPJ00605.1 KPJ09142.1 OWR42778.1 GDKB01000026 JAP38470.1 DS497669 EFA12935.1 KF768730 AIG51924.1 GEZM01016690 JAV91188.1 AAAB01008960 EAA11672.5 QOIP01000009 RLU18508.1 ADTU01014325 ADTU01014326 ADTU01014327 ADTU01014328 ADTU01014329 ADTU01014330 ADTU01014331 ADTU01014332 ADTU01014333 ADTU01014334 ACPB03000938 ACPB03000939 ACPB03000940 ACPB03000941 ACPB03000942 ACPB03000943 ACPB03000944 ACPB03000945 ACPB03000946 ACPB03000947 KQ981410 KYN41902.1 GBYB01005332 JAG75099.1 CVRI01000075 CRL08584.1 CH933809 EDW18397.1 CH940647 EDW69683.2 CH916366 EDV96485.1 CH902618 EDV41192.1 CH379070 KRT08845.1 CH954178 EDV51160.1 OUUW01000012 SPP87596.1 UFQT01000066 SSX19329.1 CH964062 EDW78721.1 EAL30350.1 CM000159 EDW93657.1 CM000363 CM002912 EDX09466.1 KMY97953.1 M97192 AE014296 BT003218 BT050435 KX016777 APF30026.1 CP012525 ALC43237.1 GL732605 EFX71861.1 GDIP01029649 JAM74066.1 GDIP01209509 JAJ13893.1 GDIP01180016 JAJ43386.1 GDIP01174566 JAJ48836.1 GDIQ01008113 JAN86624.1 KX016772 APF30021.1 GDIQ01241334 JAK10391.1 GBYB01011575 JAG81342.1 GDIP01146122 JAJ77280.1 GDIP01184715 JAJ38687.1 KQ982182 KYQ59168.1 GDIP01183366 JAJ40036.1 GDIP01163482 JAJ59920.1 GFXV01000473 MBW12278.1 KPJ00608.1 GDIP01197266 JAJ26136.1 GDIP01209510 JAJ13892.1 GDIQ01073743 GDIQ01052945 JAN20994.1 GDIQ01230429 JAK21296.1 GDIP01180017 JAJ43385.1 DS235817 EEB17589.1 GDIP01156921 JAJ66481.1 GDIP01174565 JAJ48837.1 GDIQ01021126 JAN73611.1 GDIQ01211392 JAK40333.1 GDIQ01152859 JAK98866.1 GDIP01222360 JAJ01042.1 GDIQ01076542 JAN18195.1 GDIQ01202352 JAK49373.1 GGMR01017110 MBY29729.1 GDIP01184716 JAJ38686.1 KQ436160 KOX67415.1 GDIP01183367 JAJ40035.1 CH477476 EAT40294.1 KQ414627 KOC67420.1 KQ434795 KZC05526.1 GL447910 EFN85705.1 GDIP01222361 JAJ01041.1 CH479217 EDW33705.1 KQ976514 KYM82208.1 KQ979440 KYN21485.1 HACA01001900 CDW19261.1 GDIP01138773 JAL64941.1 GL437123 EFN70991.1 GDIP01235911 JAI87490.1 KK107199 EZA55532.1 CAEY01001120 ALC43621.1 NWSH01007937 PCG62745.1 GL888365 EGI62137.1 UFQT01002179 SSX32863.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000075884
UP000007266
+ More
UP000007062 UP000279307 UP000005205 UP000015103 UP000078541 UP000183832 UP000009192 UP000008792 UP000192221 UP000001070 UP000007801 UP000001819 UP000008711 UP000268350 UP000007798 UP000002282 UP000000304 UP000000803 UP000092553 UP000000305 UP000075809 UP000009046 UP000075882 UP000075880 UP000053105 UP000008820 UP000053825 UP000076502 UP000008237 UP000008744 UP000078540 UP000005203 UP000078492 UP000000311 UP000053097 UP000015104 UP000218220 UP000007755
UP000007062 UP000279307 UP000005205 UP000015103 UP000078541 UP000183832 UP000009192 UP000008792 UP000192221 UP000001070 UP000007801 UP000001819 UP000008711 UP000268350 UP000007798 UP000002282 UP000000304 UP000000803 UP000092553 UP000000305 UP000075809 UP000009046 UP000075882 UP000075880 UP000053105 UP000008820 UP000053825 UP000076502 UP000008237 UP000008744 UP000078540 UP000005203 UP000078492 UP000000311 UP000053097 UP000015104 UP000218220 UP000007755
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
H9ITR5
A0A0V0J107
A0A2W1BAM5
A0A212EMP7
A0A2H1VKE5
A0A194QVZ8
+ More
A0A194Q6H5 A0A194QUJ7 A0A212EMS3 A0A0V0J2T4 A0A2Y9D1L1 D7EHZ7 A0A1S4G7Q1 A0A075T6Z5 A0A1Y1N186 Q7PNT0 A0A3L8DF15 A0A158NFE2 T1HDK9 A0A151JZ81 A0A0C9QXE8 A0A1J1J876 B4KVF4 B4LIB6 A0A1W4UXQ3 B4IXW4 B3M9H9 A0A0R3P8X0 B3NGJ4 A0A3B0KNZ8 A0A336LRY5 B4N2Y2 Q29DR4 B4PJQ8 B4QJS5 Q03445 A0A1L2YXN2 A0A0M4EXY0 E9H8R8 A0A0P6AXH5 A0A0P4ZYN3 A0A0P5BVK8 A0A0P5C8P6 A0A0P6IP12 A0A1L2YXM6 A0A0P5GK06 A0A0C9RSE8 A0A0P5EM22 A0A0P5BI92 A0A151XFV4 A0A0P5BKB1 A0A0P5CZ00 A0A2H8TG59 A0A194Q4Z3 A0A0P5ALF8 A0A0P4ZRV5 A0A0P6DQ48 A0A0P5HB37 A0A0N8A5P0 E0VW33 A0A0N8ADX7 A0A0P5CJN3 A0A0P6HD12 A0A0P5IK63 A0A0N8BR75 A0A0P4YWX8 A0A0P6DJM2 A0A182KPE3 A0A0P5JIQ7 A0A2S2PJV1 A0A182JJ78 A0A0P5BHC1 A0A0M8ZMS8 A0A0P5BKU0 Q170E0 A0A0L7R971 A0A154P0S2 E2BF27 A0A0N7ZQJ7 B4H799 A0A195BCD1 A0A087ZQD0 A0A195E8E3 A0A0K2T188 A0A0P5SK38 E2A6A6 A0A0P4XW61 A0A026WHJ2 T1JZC1 A0A0M4EDF2 A0A2A4ISK2 F4WUH2 A0A336MVV7
A0A194Q6H5 A0A194QUJ7 A0A212EMS3 A0A0V0J2T4 A0A2Y9D1L1 D7EHZ7 A0A1S4G7Q1 A0A075T6Z5 A0A1Y1N186 Q7PNT0 A0A3L8DF15 A0A158NFE2 T1HDK9 A0A151JZ81 A0A0C9QXE8 A0A1J1J876 B4KVF4 B4LIB6 A0A1W4UXQ3 B4IXW4 B3M9H9 A0A0R3P8X0 B3NGJ4 A0A3B0KNZ8 A0A336LRY5 B4N2Y2 Q29DR4 B4PJQ8 B4QJS5 Q03445 A0A1L2YXN2 A0A0M4EXY0 E9H8R8 A0A0P6AXH5 A0A0P4ZYN3 A0A0P5BVK8 A0A0P5C8P6 A0A0P6IP12 A0A1L2YXM6 A0A0P5GK06 A0A0C9RSE8 A0A0P5EM22 A0A0P5BI92 A0A151XFV4 A0A0P5BKB1 A0A0P5CZ00 A0A2H8TG59 A0A194Q4Z3 A0A0P5ALF8 A0A0P4ZRV5 A0A0P6DQ48 A0A0P5HB37 A0A0N8A5P0 E0VW33 A0A0N8ADX7 A0A0P5CJN3 A0A0P6HD12 A0A0P5IK63 A0A0N8BR75 A0A0P4YWX8 A0A0P6DJM2 A0A182KPE3 A0A0P5JIQ7 A0A2S2PJV1 A0A182JJ78 A0A0P5BHC1 A0A0M8ZMS8 A0A0P5BKU0 Q170E0 A0A0L7R971 A0A154P0S2 E2BF27 A0A0N7ZQJ7 B4H799 A0A195BCD1 A0A087ZQD0 A0A195E8E3 A0A0K2T188 A0A0P5SK38 E2A6A6 A0A0P4XW61 A0A026WHJ2 T1JZC1 A0A0M4EDF2 A0A2A4ISK2 F4WUH2 A0A336MVV7
PDB
5IDF
E-value=1.6099e-109,
Score=1016
Ontologies
KEGG
GO
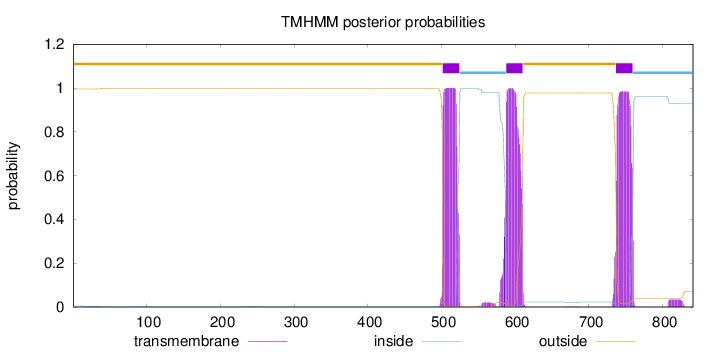
Topology
Subcellular location
Cell membrane
Cell junction
Synapse
Postsynaptic cell membrane
Length:
841
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.6874800000002
Exp number, first 60 AAs:
0.05845
Total prob of N-in:
0.00486
outside
1 - 501
TMhelix
502 - 524
inside
525 - 587
TMhelix
588 - 610
outside
611 - 736
TMhelix
737 - 759
inside
760 - 841
Population Genetic Test Statistics
Pi
3.985384
Theta
17.647859
Tajima's D
-2.007148
CLR
0.015592
CSRT
0.0126493675316234
Interpretation
Uncertain
