Gene
KWMTBOMO00229
Pre Gene Modal
BGIBMGA000643
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_UDP-N-acetylglucosamine--peptide_N-acetylglucosaminyltransferase_110_kDa_subunit-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.336 PlasmaMembrane Reliability : 1.02
Sequence
CDS
ATGGATTATCAATGCATCGTCGTCTACACCGTCATCAATATCGCTCCACCGCGTCTTACAGCTCAAGCCTCCGAATTCTCAAACGGCTGTGTCACCTGGTTCAAGAATAAAATTAAAAAAGCTCTTGCAGTGTATCTTCATTCTCTGAAATTAAATCCAAGCAACGGAATCGTGCACGGTAACTTAGCGTGTCTCTATTACAAACAAGGCTTCATAGACCTAGCCATCGATACATACAAGCGCGCTATAGAACTACAGCCTGACTTTCCGGACGCGTACTGCAATCTAGCTAATGCACTCAAAGAGAAGGGCCTTGTGACCGAAGCAGAGGAGTGCTATTTCAAAGCACTAAGTCTATGTCCCACACATGTAGATACACTCAATAATCTTGGCAACGTCAAAAGGGAACAAGGGAAAATTGATGAAGCTACAAATTTATACATGAAAGCCTTGGAGGTGTTCCCTAATTTTGCGGCAACTCATAGCAATTTGGCATCCCTTTTACAGCAACAAGGTAAACTACATCAAGCTCTATTTCACTATAAACAGGCTATCGGTATACAACCTAAGTTCGCAGATGCTTACAGTAATATGGGTAACACTCTTAGAGAGCTGCAAGACATGACGGGAGCACTTGCATGTTTCAAAAAAGCTATTGAAATAAATCCTACATTTTCTGATGCACATTGTAATTTAGCCAGCATTTACAAAGATACTGGCAACATCAAAGAAGCTATTGAGTCATACAAAAACGCTCTGTATTTTAAACCTGATTTTCCCGATGCTTATTGCAATTTAGCGCACTGTTTACAAATTGTTTGCAACTGGGATGACTACTACGAACGAATGCATAATATTATAGAAATAGTCAATAAACAATTAAAAATGGATAAACTTTCTTCTGTGCATCCACACCATTCCATCTTATATCCACTTACGAATGAAGCCAGGCGAGAAATTGCCACTCGACATGCAAATTTGTATATAGACAAACTTCATTTTTTGCAAAATACTGTCACGTTTCAACACCCCAAAGTTATGGAAGGTCGATTACGAATTGGTTACGTAAGCAGTGATTTCGGTAATCATCCAACCTCACATTTAATGCAGTCCATTCCGGGTAGCCATGATCGTTCCAAAGTGGAAGTTTTTTGTTATGCACTCAACGCTGATGATGGTACAACATTCCGTAGAAAAATAGTTGAAGAATCTGAAAATTTTATCGATTTATCTTGTATCCCTTGCAATATGGAAGCGGCTACAAGAATCCACAGAGATGGTATCCATATCTTAATTAACATGAACGGTTATACAAAGGGAGCTAGAAACGAAATTTTTGCTTTAAAACCAGCGCCTATTCAGGTTATGTGGTTGGGATATCCGGGGACAAGTGGAGCGGGCTACATAGATTATATGATAACTGATGAAGTTTCCGCACCTTTGTCGTTATCAGAAGATTTTAGTGAAAAATTTGCATATATGCCGCATACTTATTTTGTTGGTGATCACAAAAAAATGTTTCCTCACTTAAAAACGAAGTACAAGGTACTGATTGATAATAATTCGATAGCTCATGAAAATGTAGCACTGATTAATTCGTCTGAATTTATTAATATTGATGAAGTTTTTAATGTAAGATGTGATAAGACAATTGTATCCTTCGAAAATCTCGAAGAAATTGAATTTATTATAAGAAAAGTAAACATTCCGAATTATGTACTCGATACGACAATTAATCCTAAGCAAGAAAAGGTCTTAGTCAACGGTACTGTCATTGATAATGGTATATCAATAAATTATTCCAGTAAGAGAGTTGCATCCGGTGAAGTTTCTTTTGAAAATATTGTTTTTACGTCCAGAAAGCAGTACGGCTTACCTGAAGATGCAGTTGTTTTTTGCAATTTCAATCAGCTATACAAAATTGATCCAAGAATAATGGAATCTTGGGTAACTATTCTGAAACTGGTCCCTAATAGCGTACTGTGGTTGTTAAGTTTTCCTGCTGCTGGAGAACCTAATATTCAGAAATATGGGCTAAATTTAGGTTTAACACCAGGACGTATTATTTTCTCTAAAATTGCTTGTAAAGAAGAACACGTAAGACGTGGACAGTTAGCAGACATCTGCTTAGATACACCACTTTGCAACGGCCACACAACGACGATGGACATCCTCTGGACTGGCACTCCAGTGGTTACTCTGCCCGGAGATACGCTCGCTTCAAGAGTTGCTGCATCCCAGCTAACAGCACTTCATTGTACGGAGCTCATTGCTAAAAATAGAAAGCATTACGAAGATATAGCCATCAAATTAGGAACTGATTCTGCATACCGGCGATATATTCGTGCGAAAGTTTCGAAAGCTCGTTTAGAAAGCACGTTGTTTGACTGCGAGCATTATGCAAGAGGCTTGGAGAGCCTTTACTCTAAAATGTGGGAACTATATCAAAGAGGAGACAAGCCTGATCACATCGCTGTTTCTTCGAAGTGA
Protein
MDYQCIVVYTVINIAPPRLTAQASEFSNGCVTWFKNKIKKALAVYLHSLKLNPSNGIVHGNLACLYYKQGFIDLAIDTYKRAIELQPDFPDAYCNLANALKEKGLVTEAEECYFKALSLCPTHVDTLNNLGNVKREQGKIDEATNLYMKALEVFPNFAATHSNLASLLQQQGKLHQALFHYKQAIGIQPKFADAYSNMGNTLRELQDMTGALACFKKAIEINPTFSDAHCNLASIYKDTGNIKEAIESYKNALYFKPDFPDAYCNLAHCLQIVCNWDDYYERMHNIIEIVNKQLKMDKLSSVHPHHSILYPLTNEARREIATRHANLYIDKLHFLQNTVTFQHPKVMEGRLRIGYVSSDFGNHPTSHLMQSIPGSHDRSKVEVFCYALNADDGTTFRRKIVEESENFIDLSCIPCNMEAATRIHRDGIHILINMNGYTKGARNEIFALKPAPIQVMWLGYPGTSGAGYIDYMITDEVSAPLSLSEDFSEKFAYMPHTYFVGDHKKMFPHLKTKYKVLIDNNSIAHENVALINSSEFINIDEVFNVRCDKTIVSFENLEEIEFIIRKVNIPNYVLDTTINPKQEKVLVNGTVIDNGISINYSSKRVASGEVSFENIVFTSRKQYGLPEDAVVFCNFNQLYKIDPRIMESWVTILKLVPNSVLWLLSFPAAGEPNIQKYGLNLGLTPGRIIFSKIACKEEHVRRGQLADICLDTPLCNGHTTTMDILWTGTPVVTLPGDTLASRVAASQLTALHCTELIAKNRKHYEDIAIKLGTDSAYRRYIRAKVSKARLESTLFDCEHYARGLESLYSKMWELYQRGDKPDHIAVSSK
Summary
Uniprot
A0A2H1VFQ3
A0A2W1BQL6
A0A2A4JLS9
A0A212EMT0
A0A194QW03
A0A195DUJ0
+ More
A0A1S4G349 A0A088AU58 A0A182K903 A0A182PJX4 A0A182WCJ0 A0A084VDH7 A0A182RDG3 A0A182M3Z4 B0WCA4 A0A232EPS2 A0A182F9R4 A0A182NH31 W5JST6 A0A182X8M8 A0A182VB46 A0A182I146 A0A2J7PU97 A0A1W4WZX6 A0A0K8T681 A0A1B6I6U2 A0A195CB35 A0A151X021 A0A1B6C049 A0A195EWP2 A0A151HYK7 A0A158NSR3 F4W877 A0A2H1VIU1 A0A0L7QSX2 A0A2A3EG57 A0A2A4JXZ0 V9I8Q6 V9IAZ6 A0A026WWW3 A0A3L8D9Q0 Q16FI6 A0A182YPW9 A0A310SJY3 A0A2P8YNK5 A0A0M9A6T3 H9JND0 A0A194R9U0 A0A0P4VNF7 A0A182QWV3 A0A182J0E2 D6WHK3 A0A2M4CTZ1 A0A2M4A5I3 A0A2M4BB22 A0A139WK52 A0A2M4CV31 A0A2M4A4P0 A0A2M4BBC0 A0A194PPU3 A0A2M4BBG1 A0A212ETF9 A0A2M4A6B5 A0A1Q3FUS6 A0A023F5R9 A0A2M3YZF6 A0A2M4BDK8 A0A0V0G3N5 A0A182LLH0 A0A2M4BDR0 Q7Q5R0 A0A0T6B1V8 E2AQU9 A0A182U314 E0W1P3 A0A0N7ZKL5 A0A069DZQ2 A0A0P5PPV1 A0A0P4Y2T9 A0A1L8E217 Q7KJA9 A0A0A9Z4Y3 A0A0A9X5K8 A0A1Y1N3F7 A0A1B6JPC7 A0A0J9R6G6 W8AXT6 A0A182GKN8 A0A1L8E1M7 A0A0P6GV95 A0A1Y1N648 A0A1L8E1I7 A0A0Q5WP95 A0A0P5NMA8 A0A0J9S0F9 A0A3B0K6S0 A0A336MKK1 B4HCC1 B4LP13 A0A0R1E7G4
A0A1S4G349 A0A088AU58 A0A182K903 A0A182PJX4 A0A182WCJ0 A0A084VDH7 A0A182RDG3 A0A182M3Z4 B0WCA4 A0A232EPS2 A0A182F9R4 A0A182NH31 W5JST6 A0A182X8M8 A0A182VB46 A0A182I146 A0A2J7PU97 A0A1W4WZX6 A0A0K8T681 A0A1B6I6U2 A0A195CB35 A0A151X021 A0A1B6C049 A0A195EWP2 A0A151HYK7 A0A158NSR3 F4W877 A0A2H1VIU1 A0A0L7QSX2 A0A2A3EG57 A0A2A4JXZ0 V9I8Q6 V9IAZ6 A0A026WWW3 A0A3L8D9Q0 Q16FI6 A0A182YPW9 A0A310SJY3 A0A2P8YNK5 A0A0M9A6T3 H9JND0 A0A194R9U0 A0A0P4VNF7 A0A182QWV3 A0A182J0E2 D6WHK3 A0A2M4CTZ1 A0A2M4A5I3 A0A2M4BB22 A0A139WK52 A0A2M4CV31 A0A2M4A4P0 A0A2M4BBC0 A0A194PPU3 A0A2M4BBG1 A0A212ETF9 A0A2M4A6B5 A0A1Q3FUS6 A0A023F5R9 A0A2M3YZF6 A0A2M4BDK8 A0A0V0G3N5 A0A182LLH0 A0A2M4BDR0 Q7Q5R0 A0A0T6B1V8 E2AQU9 A0A182U314 E0W1P3 A0A0N7ZKL5 A0A069DZQ2 A0A0P5PPV1 A0A0P4Y2T9 A0A1L8E217 Q7KJA9 A0A0A9Z4Y3 A0A0A9X5K8 A0A1Y1N3F7 A0A1B6JPC7 A0A0J9R6G6 W8AXT6 A0A182GKN8 A0A1L8E1M7 A0A0P6GV95 A0A1Y1N648 A0A1L8E1I7 A0A0Q5WP95 A0A0P5NMA8 A0A0J9S0F9 A0A3B0K6S0 A0A336MKK1 B4HCC1 B4LP13 A0A0R1E7G4
Pubmed
28756777
22118469
26354079
17510324
24438588
28648823
+ More
20920257 23761445 21347285 21719571 24508170 30249741 25244985 29403074 19121390 27129103 18362917 19820115 25474469 20966253 12364791 14747013 17210077 20798317 20566863 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26674417 25401762 26823975 28004739 22936249 24495485 26483478 17994087 18057021 17550304
20920257 23761445 21347285 21719571 24508170 30249741 25244985 29403074 19121390 27129103 18362917 19820115 25474469 20966253 12364791 14747013 17210077 20798317 20566863 26334808 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26674417 25401762 26823975 28004739 22936249 24495485 26483478 17994087 18057021 17550304
EMBL
ODYU01002299
SOQ39611.1
KZ149977
PZC75885.1
NWSH01001075
PCG72736.1
+ More
AGBW02013810 OWR42776.1 KQ461108 KPJ09145.1 KQ980322 KYN16575.1 ATLV01011337 KE524664 KFB36021.1 AXCM01003360 DS231886 EDS43349.1 NNAY01002903 OXU20353.1 ADMH02000262 ETN67206.1 APCN01000030 NEVH01021207 PNF19894.1 GBRD01010100 GBRD01004885 GBRD01004884 JAG60936.1 GECU01025096 JAS82610.1 KQ978023 KYM98017.1 KQ982624 KYQ53682.1 GEDC01030693 GEDC01027732 JAS06605.1 JAS09566.1 KQ981948 KYN32648.1 KQ976723 KYM76681.1 ADTU01025121 GL887898 EGI69572.1 ODYU01002802 SOQ40748.1 KQ414758 KOC61571.1 KZ288255 PBC30698.1 NWSH01000423 PCG76494.1 JR036952 AEY57498.1 JR036953 AEY57499.1 KK107078 EZA60211.1 QOIP01000011 RLU16638.1 CH478429 EAT32999.1 KQ760514 OAD60022.1 PYGN01000467 PSN45840.1 KQ435724 KOX78410.1 BABH01019450 KQ460473 KPJ14407.1 GDKW01000423 JAI56172.1 AXCN02000346 KQ971331 EFA01003.1 GGFL01004463 MBW68641.1 GGFK01002651 MBW35972.1 GGFJ01001095 MBW50236.1 KYB28293.1 GGFL01005015 MBW69193.1 GGFK01002408 MBW35729.1 GGFJ01001195 MBW50336.1 KQ459597 KPI94754.1 GGFJ01001238 MBW50379.1 AGBW02012603 OWR44731.1 GGFK01003023 MBW36344.1 GFDL01003711 JAV31334.1 GBBI01002194 JAC16518.1 GGFM01000894 MBW21645.1 GGFJ01001956 MBW51097.1 GECL01003455 JAP02669.1 GGFJ01001960 MBW51101.1 AAAB01008960 EAA10760.4 LJIG01016195 KRT81330.1 GL441834 EFN64211.1 DS235873 EEB19625.1 GDIP01236217 JAI87184.1 GBGD01000245 JAC88644.1 GDIQ01221585 GDIQ01142216 GDIQ01142215 GDIQ01125129 GDIQ01113418 GDIQ01096321 GDIP01055887 GDIQ01082945 JAL26597.1 JAM47828.1 GDIP01234374 JAI89027.1 GFDF01001462 JAV12622.1 AF217788 AE013599 BT032865 AAF32311.1 AAF57338.1 AAG22338.1 AAG22339.1 ACD81879.1 GBHO01003282 GDHC01006945 JAG40322.1 JAQ11684.1 GBHO01027602 JAG16002.1 GEZM01013689 GEZM01013687 JAV92369.1 GECU01006701 JAT01006.1 CM002911 KMY91610.1 KMY91611.1 GAMC01012750 JAB93805.1 JXUM01071183 JXUM01071184 KQ562648 KXJ75410.1 GFDF01001473 JAV12611.1 GDIQ01041005 JAN53732.1 GEZM01013688 JAV92370.1 GFDF01001494 JAV12590.1 CH954177 KQS71055.1 GDIQ01140365 JAL11361.1 CM002912 KMZ01359.1 OUUW01000032 SPP89814.1 UFQT01001415 SSX30450.1 CH479296 EDW25761.1 CH940648 EDW61182.1 KRF79836.1 KRF79837.1 CH891608 KRK05133.1
AGBW02013810 OWR42776.1 KQ461108 KPJ09145.1 KQ980322 KYN16575.1 ATLV01011337 KE524664 KFB36021.1 AXCM01003360 DS231886 EDS43349.1 NNAY01002903 OXU20353.1 ADMH02000262 ETN67206.1 APCN01000030 NEVH01021207 PNF19894.1 GBRD01010100 GBRD01004885 GBRD01004884 JAG60936.1 GECU01025096 JAS82610.1 KQ978023 KYM98017.1 KQ982624 KYQ53682.1 GEDC01030693 GEDC01027732 JAS06605.1 JAS09566.1 KQ981948 KYN32648.1 KQ976723 KYM76681.1 ADTU01025121 GL887898 EGI69572.1 ODYU01002802 SOQ40748.1 KQ414758 KOC61571.1 KZ288255 PBC30698.1 NWSH01000423 PCG76494.1 JR036952 AEY57498.1 JR036953 AEY57499.1 KK107078 EZA60211.1 QOIP01000011 RLU16638.1 CH478429 EAT32999.1 KQ760514 OAD60022.1 PYGN01000467 PSN45840.1 KQ435724 KOX78410.1 BABH01019450 KQ460473 KPJ14407.1 GDKW01000423 JAI56172.1 AXCN02000346 KQ971331 EFA01003.1 GGFL01004463 MBW68641.1 GGFK01002651 MBW35972.1 GGFJ01001095 MBW50236.1 KYB28293.1 GGFL01005015 MBW69193.1 GGFK01002408 MBW35729.1 GGFJ01001195 MBW50336.1 KQ459597 KPI94754.1 GGFJ01001238 MBW50379.1 AGBW02012603 OWR44731.1 GGFK01003023 MBW36344.1 GFDL01003711 JAV31334.1 GBBI01002194 JAC16518.1 GGFM01000894 MBW21645.1 GGFJ01001956 MBW51097.1 GECL01003455 JAP02669.1 GGFJ01001960 MBW51101.1 AAAB01008960 EAA10760.4 LJIG01016195 KRT81330.1 GL441834 EFN64211.1 DS235873 EEB19625.1 GDIP01236217 JAI87184.1 GBGD01000245 JAC88644.1 GDIQ01221585 GDIQ01142216 GDIQ01142215 GDIQ01125129 GDIQ01113418 GDIQ01096321 GDIP01055887 GDIQ01082945 JAL26597.1 JAM47828.1 GDIP01234374 JAI89027.1 GFDF01001462 JAV12622.1 AF217788 AE013599 BT032865 AAF32311.1 AAF57338.1 AAG22338.1 AAG22339.1 ACD81879.1 GBHO01003282 GDHC01006945 JAG40322.1 JAQ11684.1 GBHO01027602 JAG16002.1 GEZM01013689 GEZM01013687 JAV92369.1 GECU01006701 JAT01006.1 CM002911 KMY91610.1 KMY91611.1 GAMC01012750 JAB93805.1 JXUM01071183 JXUM01071184 KQ562648 KXJ75410.1 GFDF01001473 JAV12611.1 GDIQ01041005 JAN53732.1 GEZM01013688 JAV92370.1 GFDF01001494 JAV12590.1 CH954177 KQS71055.1 GDIQ01140365 JAL11361.1 CM002912 KMZ01359.1 OUUW01000032 SPP89814.1 UFQT01001415 SSX30450.1 CH479296 EDW25761.1 CH940648 EDW61182.1 KRF79836.1 KRF79837.1 CH891608 KRK05133.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000078492
UP000005203
UP000075881
+ More
UP000075885 UP000075920 UP000030765 UP000075900 UP000075883 UP000002320 UP000215335 UP000069272 UP000075884 UP000000673 UP000076407 UP000075903 UP000075840 UP000235965 UP000192223 UP000078542 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000053825 UP000242457 UP000053097 UP000279307 UP000008820 UP000076408 UP000245037 UP000053105 UP000005204 UP000075886 UP000075880 UP000007266 UP000053268 UP000075882 UP000007062 UP000000311 UP000075902 UP000009046 UP000000803 UP000069940 UP000249989 UP000008711 UP000268350 UP000008744 UP000008792 UP000002282
UP000075885 UP000075920 UP000030765 UP000075900 UP000075883 UP000002320 UP000215335 UP000069272 UP000075884 UP000000673 UP000076407 UP000075903 UP000075840 UP000235965 UP000192223 UP000078542 UP000075809 UP000078541 UP000078540 UP000005205 UP000007755 UP000053825 UP000242457 UP000053097 UP000279307 UP000008820 UP000076408 UP000245037 UP000053105 UP000005204 UP000075886 UP000075880 UP000007266 UP000053268 UP000075882 UP000007062 UP000000311 UP000075902 UP000009046 UP000000803 UP000069940 UP000249989 UP000008711 UP000268350 UP000008744 UP000008792 UP000002282
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VFQ3
A0A2W1BQL6
A0A2A4JLS9
A0A212EMT0
A0A194QW03
A0A195DUJ0
+ More
A0A1S4G349 A0A088AU58 A0A182K903 A0A182PJX4 A0A182WCJ0 A0A084VDH7 A0A182RDG3 A0A182M3Z4 B0WCA4 A0A232EPS2 A0A182F9R4 A0A182NH31 W5JST6 A0A182X8M8 A0A182VB46 A0A182I146 A0A2J7PU97 A0A1W4WZX6 A0A0K8T681 A0A1B6I6U2 A0A195CB35 A0A151X021 A0A1B6C049 A0A195EWP2 A0A151HYK7 A0A158NSR3 F4W877 A0A2H1VIU1 A0A0L7QSX2 A0A2A3EG57 A0A2A4JXZ0 V9I8Q6 V9IAZ6 A0A026WWW3 A0A3L8D9Q0 Q16FI6 A0A182YPW9 A0A310SJY3 A0A2P8YNK5 A0A0M9A6T3 H9JND0 A0A194R9U0 A0A0P4VNF7 A0A182QWV3 A0A182J0E2 D6WHK3 A0A2M4CTZ1 A0A2M4A5I3 A0A2M4BB22 A0A139WK52 A0A2M4CV31 A0A2M4A4P0 A0A2M4BBC0 A0A194PPU3 A0A2M4BBG1 A0A212ETF9 A0A2M4A6B5 A0A1Q3FUS6 A0A023F5R9 A0A2M3YZF6 A0A2M4BDK8 A0A0V0G3N5 A0A182LLH0 A0A2M4BDR0 Q7Q5R0 A0A0T6B1V8 E2AQU9 A0A182U314 E0W1P3 A0A0N7ZKL5 A0A069DZQ2 A0A0P5PPV1 A0A0P4Y2T9 A0A1L8E217 Q7KJA9 A0A0A9Z4Y3 A0A0A9X5K8 A0A1Y1N3F7 A0A1B6JPC7 A0A0J9R6G6 W8AXT6 A0A182GKN8 A0A1L8E1M7 A0A0P6GV95 A0A1Y1N648 A0A1L8E1I7 A0A0Q5WP95 A0A0P5NMA8 A0A0J9S0F9 A0A3B0K6S0 A0A336MKK1 B4HCC1 B4LP13 A0A0R1E7G4
A0A1S4G349 A0A088AU58 A0A182K903 A0A182PJX4 A0A182WCJ0 A0A084VDH7 A0A182RDG3 A0A182M3Z4 B0WCA4 A0A232EPS2 A0A182F9R4 A0A182NH31 W5JST6 A0A182X8M8 A0A182VB46 A0A182I146 A0A2J7PU97 A0A1W4WZX6 A0A0K8T681 A0A1B6I6U2 A0A195CB35 A0A151X021 A0A1B6C049 A0A195EWP2 A0A151HYK7 A0A158NSR3 F4W877 A0A2H1VIU1 A0A0L7QSX2 A0A2A3EG57 A0A2A4JXZ0 V9I8Q6 V9IAZ6 A0A026WWW3 A0A3L8D9Q0 Q16FI6 A0A182YPW9 A0A310SJY3 A0A2P8YNK5 A0A0M9A6T3 H9JND0 A0A194R9U0 A0A0P4VNF7 A0A182QWV3 A0A182J0E2 D6WHK3 A0A2M4CTZ1 A0A2M4A5I3 A0A2M4BB22 A0A139WK52 A0A2M4CV31 A0A2M4A4P0 A0A2M4BBC0 A0A194PPU3 A0A2M4BBG1 A0A212ETF9 A0A2M4A6B5 A0A1Q3FUS6 A0A023F5R9 A0A2M3YZF6 A0A2M4BDK8 A0A0V0G3N5 A0A182LLH0 A0A2M4BDR0 Q7Q5R0 A0A0T6B1V8 E2AQU9 A0A182U314 E0W1P3 A0A0N7ZKL5 A0A069DZQ2 A0A0P5PPV1 A0A0P4Y2T9 A0A1L8E217 Q7KJA9 A0A0A9Z4Y3 A0A0A9X5K8 A0A1Y1N3F7 A0A1B6JPC7 A0A0J9R6G6 W8AXT6 A0A182GKN8 A0A1L8E1M7 A0A0P6GV95 A0A1Y1N648 A0A1L8E1I7 A0A0Q5WP95 A0A0P5NMA8 A0A0J9S0F9 A0A3B0K6S0 A0A336MKK1 B4HCC1 B4LP13 A0A0R1E7G4
PDB
5A01
E-value=0,
Score=2477
Ontologies
GO
PANTHER
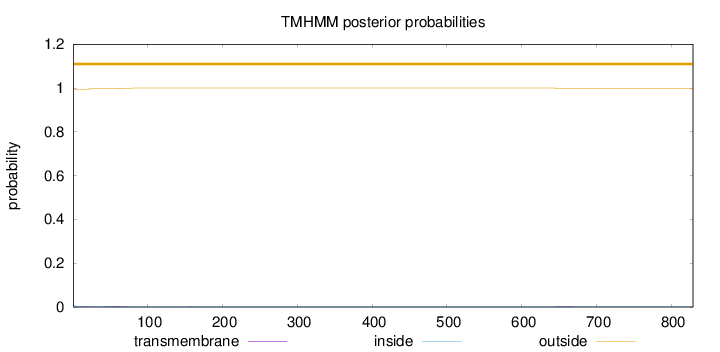
Topology
Length:
829
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.137580000000001
Exp number, first 60 AAs:
0.0912900000000001
Total prob of N-in:
0.00542
outside
1 - 829
Population Genetic Test Statistics
Pi
21.283792
Theta
22.940449
Tajima's D
0.044781
CLR
0.008134
CSRT
0.387780610969452
Interpretation
Uncertain
