Gene
KWMTBOMO00228
Pre Gene Modal
BGIBMGA000642
Annotation
tryptophan_5-hydroxylase_1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.511
Sequence
CDS
ATGAGTGGTTCGGGAAAAGGTCTTCTAGGTCTTTGGCTCTATAGAAACGGTTCTGATTGGCAAGTAAAGAATGAATCTCCACACCATCCTAAGCTCGGAGAGCTTCATGCAGCAAGAGTCCAGGCTCAAGCAGAAAGCGAACGTATTTCGGTGATGTTTACATTGAAAAACCAAGTTGGCGGACTAGTTCGGGCCTTATCCGTTTTCCAAGACTTGGGTATTAACGTTCTCCACATTGAATCCAGAAAATCTATGACTGAAGTTTCATCTGCCGACATCCTCGTGGACGTGGAGTGTGATCCCCAAAGAATGGAGCAACTGAAGAGGATGCTAAAACGAGAAGTCCAAGACTTTGAGGTTGTACCACCGCAGACTGGTGATGAGTTTCCTCCTCCAACGCCTATGTCTGCTGCGGCAAGCTTCGATTTCGGTGAGATGCCCTGGTTTCCACGAAAGATATCTGACTTGGACCGCGCCCAAAACGTCCTCATGTATGGTTCGGAACTGGACGCCGATCACCCTGGCTTCAAAGACCCGATATATAGGAAACGCCGTGAACAGTTTGCGGCAATCGCCAATAACTATAAGTATGGCCATCCGATCCCGAAGGTGCAGTATACTGAAACAGAAATCAAAACATGGGGAATTGTGTTCCGTGAGCTTCACAAACTTTACCAGAAACACGCTTGTCCTGAGTACTTAGAGAACTGGCCCCAACTTGTCAAGTACTGCGGCTACAGGGAGGACAACTTGCCTCAGCTAGAAGATGTGAGTTCATTCTTGAAACGAAAAACTGGCTTTCAATTGCGCCCTGTTGCCGGCTATCTCTCGCCGAGGGACTTTTTATCGGGTCTTGCTTTCCGTGTTTTCCACTGCACCCAATACATTCGTCACTCGTCTGATCCTTTCTACACTCCGGAACCCGATTGCTGCCATGAACTTCTCGGGCACATGCCGCTGCTAGCTAATCCAAGTTTCGCGCAATTCTCTCAAGAACTCGGCTTGGCGTCTCTCGGCGCGTCTGATGCAGACATAGATAAATTGGCTACCTTGTATTTCTTCACGGTTGAATTTGGCTTGTGTCGTCAACCGGATGGCTCTTTTTGCGTTTACGGAGCTGGACTGCTTTCGTCTGTGGCTGAACTCCAACATGCCCTCACTACGCCGGAAAAAATAAAGCGTTTCGACCCCGATATTACTGTACATGAAGAATGCATCATCACTTCATACCAAAACGCTTATTATTATACCGATTCCTTTGAAGAGGCGAAGGAGAAAATGAGAGCATTTGCTGAAAGCATACAACGCCCTTTTGGAGTCAGATACAATCCATACACTCAAAGCGTCGAGGTACTGAGTAATGCACAGAAGATTACTGCTCTGGTCTCTGAGTTACGAGGAGATTTGTGCATTGTGTCTTCGGCTATAAAGAAAATATCAGCTCAGGATTCGACATTAGATGTAGAGTCGATTGCTAATATGCTTCATACTGGTCTTCAGGTTCACGATCGGAGTCCACAGAGCACGTCTGGAGCGAGCACTCCTAATTCTGAACGCGGTACTTCGCCAGCAAGGCTAGAGAAAACCGAAGAAACTCTTAAAACCGACGAAGAAAAACCAACTAAAAACTAA
Protein
MSGSGKGLLGLWLYRNGSDWQVKNESPHHPKLGELHAARVQAQAESERISVMFTLKNQVGGLVRALSVFQDLGINVLHIESRKSMTEVSSADILVDVECDPQRMEQLKRMLKREVQDFEVVPPQTGDEFPPPTPMSAAASFDFGEMPWFPRKISDLDRAQNVLMYGSELDADHPGFKDPIYRKRREQFAAIANNYKYGHPIPKVQYTETEIKTWGIVFRELHKLYQKHACPEYLENWPQLVKYCGYREDNLPQLEDVSSFLKRKTGFQLRPVAGYLSPRDFLSGLAFRVFHCTQYIRHSSDPFYTPEPDCCHELLGHMPLLANPSFAQFSQELGLASLGASDADIDKLATLYFFTVEFGLCRQPDGSFCVYGAGLLSSVAELQHALTTPEKIKRFDPDITVHEECIITSYQNAYYYTDSFEEAKEKMRAFAESIQRPFGVRYNPYTQSVEVLSNAQKITALVSELRGDLCIVSSAIKKISAQDSTLDVESIANMLHTGLQVHDRSPQSTSGASTPNSERGTSPARLEKTEETLKTDEEKPTKN
Summary
Cofactor
Fe(2+)
Uniprot
A0A0A0QIF7
H9ITR2
A0A2H1VFH5
A0A2A4JKX5
A0A194QZX5
A0A212EMQ3
+ More
A0A0G3VJK8 A0A142F422 A0A194QB12 A0A084W0Q8 A0A182IYT9 A0A1S4G590 J9E928 A0A182I1Q9 A0A182YCW9 A0A182KBJ1 A0A182PJI9 A0A182WKJ5 A0A182FCQ7 A0A182KPE9 A0A182RPF4 A0A1J1I820 A0A182GEG7 A0A336LM66 A0A182NKC7 Q7Q665 W5JIU7 A0A182SWG0 D6WHK1 B0XK29 A0A1B0CQB7 A0A2H8TWM4 A0A1I8P648 A0A1I8MZC1 J9JJ41 E9RJU9 A0A2A3EIN8 A0A1W4X3N3 W8C3W8 A0A0K8VBY4 B4KWR4 R9S2K6 A0A034WR65 E2AD07 A0A3B0KPT3 B4MM34 A0A0R3P6T9 A0A1A9Y2A1 A0A1B0B9H9 A0A195DAI2 A0A087ZZK7 B3M555 E2BF35 Q29DP1 A0A0L7R9H6 A0A158NFE8 B4IZR0 A0A310SIY1 B4HVN4 A0A1B0AD17 A0A1W4W6M0 B4LGY7 A0A0J9RMW1 B4PCR3 A0A0L0BV79 Q9W0K2 B4QLX4 B3NJF1 A0A182ME81 A0A182QL96 A0A195BDE8 A0A067QRH3 A0A182X774 A0A151IHU0 F2ZBU1 A0A182UZZ4 A0A1A9X2W7 K7IMD2 E0VZU7 B4HDG7 T1I7M5 A0A1D2MRD4 A0A0N0BKX3 A0A226DK15 A0A1B0GCQ6 A0A0C9QF55 N6U2M6 A0A026WHP4 A0A0P5FLH1 A0A0P6AWJ1 A0A0P5YZL0 E9G1D2 T1KEU9 T1J5F7 A0A1S3JT92 A0A210QFD6 V3ZXB0
A0A0G3VJK8 A0A142F422 A0A194QB12 A0A084W0Q8 A0A182IYT9 A0A1S4G590 J9E928 A0A182I1Q9 A0A182YCW9 A0A182KBJ1 A0A182PJI9 A0A182WKJ5 A0A182FCQ7 A0A182KPE9 A0A182RPF4 A0A1J1I820 A0A182GEG7 A0A336LM66 A0A182NKC7 Q7Q665 W5JIU7 A0A182SWG0 D6WHK1 B0XK29 A0A1B0CQB7 A0A2H8TWM4 A0A1I8P648 A0A1I8MZC1 J9JJ41 E9RJU9 A0A2A3EIN8 A0A1W4X3N3 W8C3W8 A0A0K8VBY4 B4KWR4 R9S2K6 A0A034WR65 E2AD07 A0A3B0KPT3 B4MM34 A0A0R3P6T9 A0A1A9Y2A1 A0A1B0B9H9 A0A195DAI2 A0A087ZZK7 B3M555 E2BF35 Q29DP1 A0A0L7R9H6 A0A158NFE8 B4IZR0 A0A310SIY1 B4HVN4 A0A1B0AD17 A0A1W4W6M0 B4LGY7 A0A0J9RMW1 B4PCR3 A0A0L0BV79 Q9W0K2 B4QLX4 B3NJF1 A0A182ME81 A0A182QL96 A0A195BDE8 A0A067QRH3 A0A182X774 A0A151IHU0 F2ZBU1 A0A182UZZ4 A0A1A9X2W7 K7IMD2 E0VZU7 B4HDG7 T1I7M5 A0A1D2MRD4 A0A0N0BKX3 A0A226DK15 A0A1B0GCQ6 A0A0C9QF55 N6U2M6 A0A026WHP4 A0A0P5FLH1 A0A0P6AWJ1 A0A0P5YZL0 E9G1D2 T1KEU9 T1J5F7 A0A1S3JT92 A0A210QFD6 V3ZXB0
Pubmed
19121390
26354079
22118469
26974346
24438588
17510324
+ More
25244985 20966253 26483478 12364791 20920257 23761445 18362917 19820115 25315136 24495485 17994087 25348373 20798317 15632085 21347285 22936249 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 20075255 20566863 27289101 23537049 24508170 30249741 21292972 28812685 23254933
25244985 20966253 26483478 12364791 20920257 23761445 18362917 19820115 25315136 24495485 17994087 25348373 20798317 15632085 21347285 22936249 17550304 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 20075255 20566863 27289101 23537049 24508170 30249741 21292972 28812685 23254933
EMBL
KF650639
AIL94234.1
BABH01006872
ODYU01002299
SOQ39610.1
NWSH01001075
+ More
PCG72735.1 KQ461108 KPJ09146.1 AGBW02013810 OWR42775.1 KP657632 RSAL01000121 AKL78857.1 RVE46690.1 KT946792 AMQ67551.1 KQ459463 KPJ00611.1 ATLV01019151 ATLV01019152 KE525263 KFB43802.1 CH477226 EJY57396.1 APCN01000088 CVRI01000042 CRK95724.1 JXUM01059094 KQ562035 KXJ76846.1 UFQS01000067 UFQT01000067 SSW98952.1 SSX19334.1 AAAB01008960 EAA11742.3 ADMH02001054 ETN64302.1 KQ971331 EFA01002.1 DS233689 EDS31126.1 AJWK01023383 AJWK01023384 AJWK01023385 AJWK01023386 GFXV01006888 MBW18693.1 ABLF02030381 AB618095 BAJ83476.1 KZ288230 PBC31587.1 GAMC01002602 JAC03954.1 GDHF01015972 JAI36342.1 CH933809 EDW17511.1 KC699559 AGN03862.1 GAKP01000871 JAC58081.1 GL438633 EFN68680.1 OUUW01000012 SPP87201.1 CH963847 EDW73043.1 CH379070 KRT08873.1 JXJN01010387 JXJN01010388 KQ981063 KYN09920.1 CH902618 EDV39534.1 GL447910 EFN85713.1 EAL30373.2 KQ414627 KOC67406.1 ADTU01014379 ADTU01014380 CH916366 EDV95645.1 KQ767065 OAD53510.1 CH480817 EDW49999.1 CH940647 EDW68457.2 CM002912 KMY96754.1 CM000159 EDW92786.1 JRES01001279 KNC23970.1 AE014296 BT001418 AAF47444.1 AAN71173.1 CM000363 EDX08760.1 CH954178 EDV50113.1 AXCM01011779 AXCN02000407 KQ976514 KYM82227.1 KK853024 KDR12331.1 KQ977578 KYN01817.1 AB626809 BAK19479.1 DS235854 EEB18903.1 CH480298 EDW27421.1 ACPB03000182 ACPB03000183 LJIJ01000646 ODM95603.1 KQ435691 KOX81148.1 LNIX01000017 OXA45573.1 CCAG010002190 GBYB01013133 JAG82900.1 APGK01041859 KB740998 ENN75820.1 KK107199 QOIP01000009 EZA55550.1 RLU18973.1 GDIQ01259517 JAJ92207.1 GDIP01029905 LRGB01000084 JAM73810.1 KZS20986.1 GDIP01064301 JAM39414.1 GL732529 EFX86644.1 CAEY01000029 JH431861 NEDP02003883 OWF47473.1 KB202953 ESO87260.1
PCG72735.1 KQ461108 KPJ09146.1 AGBW02013810 OWR42775.1 KP657632 RSAL01000121 AKL78857.1 RVE46690.1 KT946792 AMQ67551.1 KQ459463 KPJ00611.1 ATLV01019151 ATLV01019152 KE525263 KFB43802.1 CH477226 EJY57396.1 APCN01000088 CVRI01000042 CRK95724.1 JXUM01059094 KQ562035 KXJ76846.1 UFQS01000067 UFQT01000067 SSW98952.1 SSX19334.1 AAAB01008960 EAA11742.3 ADMH02001054 ETN64302.1 KQ971331 EFA01002.1 DS233689 EDS31126.1 AJWK01023383 AJWK01023384 AJWK01023385 AJWK01023386 GFXV01006888 MBW18693.1 ABLF02030381 AB618095 BAJ83476.1 KZ288230 PBC31587.1 GAMC01002602 JAC03954.1 GDHF01015972 JAI36342.1 CH933809 EDW17511.1 KC699559 AGN03862.1 GAKP01000871 JAC58081.1 GL438633 EFN68680.1 OUUW01000012 SPP87201.1 CH963847 EDW73043.1 CH379070 KRT08873.1 JXJN01010387 JXJN01010388 KQ981063 KYN09920.1 CH902618 EDV39534.1 GL447910 EFN85713.1 EAL30373.2 KQ414627 KOC67406.1 ADTU01014379 ADTU01014380 CH916366 EDV95645.1 KQ767065 OAD53510.1 CH480817 EDW49999.1 CH940647 EDW68457.2 CM002912 KMY96754.1 CM000159 EDW92786.1 JRES01001279 KNC23970.1 AE014296 BT001418 AAF47444.1 AAN71173.1 CM000363 EDX08760.1 CH954178 EDV50113.1 AXCM01011779 AXCN02000407 KQ976514 KYM82227.1 KK853024 KDR12331.1 KQ977578 KYN01817.1 AB626809 BAK19479.1 DS235854 EEB18903.1 CH480298 EDW27421.1 ACPB03000182 ACPB03000183 LJIJ01000646 ODM95603.1 KQ435691 KOX81148.1 LNIX01000017 OXA45573.1 CCAG010002190 GBYB01013133 JAG82900.1 APGK01041859 KB740998 ENN75820.1 KK107199 QOIP01000009 EZA55550.1 RLU18973.1 GDIQ01259517 JAJ92207.1 GDIP01029905 LRGB01000084 JAM73810.1 KZS20986.1 GDIP01064301 JAM39414.1 GL732529 EFX86644.1 CAEY01000029 JH431861 NEDP02003883 OWF47473.1 KB202953 ESO87260.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000283053
UP000053268
+ More
UP000030765 UP000075880 UP000008820 UP000075840 UP000076408 UP000075881 UP000075885 UP000075920 UP000069272 UP000075882 UP000075900 UP000183832 UP000069940 UP000249989 UP000075884 UP000007062 UP000000673 UP000075901 UP000007266 UP000002320 UP000092461 UP000095300 UP000095301 UP000007819 UP000242457 UP000192223 UP000009192 UP000000311 UP000268350 UP000007798 UP000001819 UP000092443 UP000092460 UP000078492 UP000005203 UP000007801 UP000008237 UP000053825 UP000005205 UP000001070 UP000001292 UP000092445 UP000192221 UP000008792 UP000002282 UP000037069 UP000000803 UP000000304 UP000008711 UP000075883 UP000075886 UP000078540 UP000027135 UP000076407 UP000078542 UP000075903 UP000091820 UP000002358 UP000009046 UP000008744 UP000015103 UP000094527 UP000053105 UP000198287 UP000092444 UP000019118 UP000053097 UP000279307 UP000076858 UP000000305 UP000015104 UP000085678 UP000242188 UP000030746
UP000030765 UP000075880 UP000008820 UP000075840 UP000076408 UP000075881 UP000075885 UP000075920 UP000069272 UP000075882 UP000075900 UP000183832 UP000069940 UP000249989 UP000075884 UP000007062 UP000000673 UP000075901 UP000007266 UP000002320 UP000092461 UP000095300 UP000095301 UP000007819 UP000242457 UP000192223 UP000009192 UP000000311 UP000268350 UP000007798 UP000001819 UP000092443 UP000092460 UP000078492 UP000005203 UP000007801 UP000008237 UP000053825 UP000005205 UP000001070 UP000001292 UP000092445 UP000192221 UP000008792 UP000002282 UP000037069 UP000000803 UP000000304 UP000008711 UP000075883 UP000075886 UP000078540 UP000027135 UP000076407 UP000078542 UP000075903 UP000091820 UP000002358 UP000009046 UP000008744 UP000015103 UP000094527 UP000053105 UP000198287 UP000092444 UP000019118 UP000053097 UP000279307 UP000076858 UP000000305 UP000015104 UP000085678 UP000242188 UP000030746
Interpro
IPR036329
Aro-AA_hydroxylase_C_sf
+ More
IPR019773 Tyrosine_3-monooxygenase-like
IPR001273 ArAA_hydroxylase
IPR002912 ACT_dom
IPR018301 ArAA_hydroxylase_Fe/CU_BS
IPR041904 TrpOH_cat
IPR005963 Trp_5_mOase
IPR019774 Aromatic-AA_hydroxylase_C
IPR036951 ArAA_hydroxylase_sf
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR001440 TPR_1
IPR037919 OGT
IPR013026 TPR-contain_dom
IPR029489 OGT/SEC/SPY_C
IPR019773 Tyrosine_3-monooxygenase-like
IPR001273 ArAA_hydroxylase
IPR002912 ACT_dom
IPR018301 ArAA_hydroxylase_Fe/CU_BS
IPR041904 TrpOH_cat
IPR005963 Trp_5_mOase
IPR019774 Aromatic-AA_hydroxylase_C
IPR036951 ArAA_hydroxylase_sf
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR001440 TPR_1
IPR037919 OGT
IPR013026 TPR-contain_dom
IPR029489 OGT/SEC/SPY_C
Gene 3D
CDD
ProteinModelPortal
A0A0A0QIF7
H9ITR2
A0A2H1VFH5
A0A2A4JKX5
A0A194QZX5
A0A212EMQ3
+ More
A0A0G3VJK8 A0A142F422 A0A194QB12 A0A084W0Q8 A0A182IYT9 A0A1S4G590 J9E928 A0A182I1Q9 A0A182YCW9 A0A182KBJ1 A0A182PJI9 A0A182WKJ5 A0A182FCQ7 A0A182KPE9 A0A182RPF4 A0A1J1I820 A0A182GEG7 A0A336LM66 A0A182NKC7 Q7Q665 W5JIU7 A0A182SWG0 D6WHK1 B0XK29 A0A1B0CQB7 A0A2H8TWM4 A0A1I8P648 A0A1I8MZC1 J9JJ41 E9RJU9 A0A2A3EIN8 A0A1W4X3N3 W8C3W8 A0A0K8VBY4 B4KWR4 R9S2K6 A0A034WR65 E2AD07 A0A3B0KPT3 B4MM34 A0A0R3P6T9 A0A1A9Y2A1 A0A1B0B9H9 A0A195DAI2 A0A087ZZK7 B3M555 E2BF35 Q29DP1 A0A0L7R9H6 A0A158NFE8 B4IZR0 A0A310SIY1 B4HVN4 A0A1B0AD17 A0A1W4W6M0 B4LGY7 A0A0J9RMW1 B4PCR3 A0A0L0BV79 Q9W0K2 B4QLX4 B3NJF1 A0A182ME81 A0A182QL96 A0A195BDE8 A0A067QRH3 A0A182X774 A0A151IHU0 F2ZBU1 A0A182UZZ4 A0A1A9X2W7 K7IMD2 E0VZU7 B4HDG7 T1I7M5 A0A1D2MRD4 A0A0N0BKX3 A0A226DK15 A0A1B0GCQ6 A0A0C9QF55 N6U2M6 A0A026WHP4 A0A0P5FLH1 A0A0P6AWJ1 A0A0P5YZL0 E9G1D2 T1KEU9 T1J5F7 A0A1S3JT92 A0A210QFD6 V3ZXB0
A0A0G3VJK8 A0A142F422 A0A194QB12 A0A084W0Q8 A0A182IYT9 A0A1S4G590 J9E928 A0A182I1Q9 A0A182YCW9 A0A182KBJ1 A0A182PJI9 A0A182WKJ5 A0A182FCQ7 A0A182KPE9 A0A182RPF4 A0A1J1I820 A0A182GEG7 A0A336LM66 A0A182NKC7 Q7Q665 W5JIU7 A0A182SWG0 D6WHK1 B0XK29 A0A1B0CQB7 A0A2H8TWM4 A0A1I8P648 A0A1I8MZC1 J9JJ41 E9RJU9 A0A2A3EIN8 A0A1W4X3N3 W8C3W8 A0A0K8VBY4 B4KWR4 R9S2K6 A0A034WR65 E2AD07 A0A3B0KPT3 B4MM34 A0A0R3P6T9 A0A1A9Y2A1 A0A1B0B9H9 A0A195DAI2 A0A087ZZK7 B3M555 E2BF35 Q29DP1 A0A0L7R9H6 A0A158NFE8 B4IZR0 A0A310SIY1 B4HVN4 A0A1B0AD17 A0A1W4W6M0 B4LGY7 A0A0J9RMW1 B4PCR3 A0A0L0BV79 Q9W0K2 B4QLX4 B3NJF1 A0A182ME81 A0A182QL96 A0A195BDE8 A0A067QRH3 A0A182X774 A0A151IHU0 F2ZBU1 A0A182UZZ4 A0A1A9X2W7 K7IMD2 E0VZU7 B4HDG7 T1I7M5 A0A1D2MRD4 A0A0N0BKX3 A0A226DK15 A0A1B0GCQ6 A0A0C9QF55 N6U2M6 A0A026WHP4 A0A0P5FLH1 A0A0P6AWJ1 A0A0P5YZL0 E9G1D2 T1KEU9 T1J5F7 A0A1S3JT92 A0A210QFD6 V3ZXB0
PDB
5L01
E-value=3.12103e-159,
Score=1443
Ontologies
KEGG
PATHWAY
GO
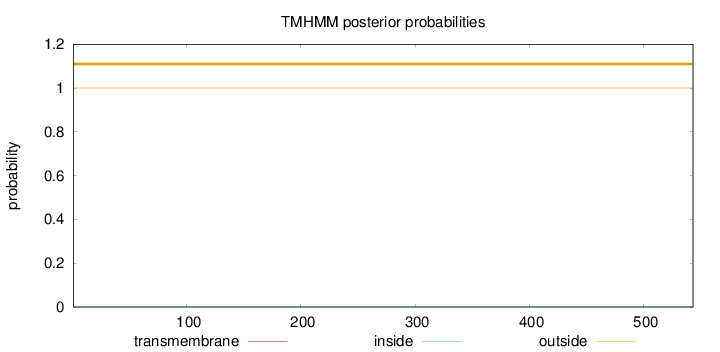
Topology
Length:
543
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00262
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00025
outside
1 - 543
Population Genetic Test Statistics
Pi
19.510695
Theta
22.287126
Tajima's D
-1.190263
CLR
0.098028
CSRT
0.106244687765612
Interpretation
Uncertain
