Gene
KWMTBOMO00227
Pre Gene Modal
BGIBMGA000570
Annotation
PREDICTED:_JNK-interacting_protein_1_[Plutella_xylostella]
Full name
JNK-interacting protein 1
Alternative Name
APP-like-interacting protein 1
Protein eye developmental SP512
Protein eye developmental SP512
Location in the cell
Nuclear Reliability : 3.849
Sequence
CDS
ATGGCCGATAGCGAGTTTGAAGAATTTCGACGAGTTTTTGATAGACTACCGCAACACATTAAGGCCCCAACGCCTTTTTACTCATTAGTGCCAAATGTTGGATTCGAAGATGAGTCACCTTCCTCGAAAAGTGAAGGTAGCCCCGAAGAAGACAGAGGATGTGTAGGCGAAACCGATGGTCGGTCATCTATTCCGGAAAATCCGGAGTCTCCACCGCCACCGGCAGACGCAGCAGACACACAACGCAGTCCTCGTCACCAGACGTTTACAAATGGCGAAAGGCGAAGGAGAAAACTCCCTGAGATCCCGAAACATAAAAAATCATCTATTCTAGCATGTGGCCAACCAGCATCTCTAGCTGATGAACTTGGGGCTCTAACCGGGATGCTCGTAAGACCAGGATGTCACAGCAGACCACTGCTCGTGCTAAAATGTGGCTATCTCCTCGACGAAGACTCCAGCCCAGATTCGGAGAGACTGCAAAGCTTGGGAGATGTCGACAGTGGACACAGCACCGCACATTCCCCTATCGACACGGGTCCAAAGAGTTTGTCGCCAATACCCTTACAGCAAAACGTGTCGCCCTCGTCCAGTTGCAGTATCAGTGGAATGAATTTTGCAGGAAATATGACCACAGTACCTCATAGTCAACTAGAAATGCTTGAAGCAACCCACAGAGGCCTACATAAATTTAACCCTAGGCATCACGATGAAATCGAGGTGGAAATAGGGGACCCTATTTATGTTCAAAAAGAAGCAGAAGATCTATGGTGCGAAGGTGTAAATTTACGAACAGGACAGCAAGGAATCTTTCCCTCCGCCTACGCAGTCGATATGGATTACAACGACTTCGACCCAGCAAATGCCAAAGTAAAAAGGGAACGATATTTATTAGGGTATATGGGCTCCGTAGAAACTTTAGCCCATAAAGGTACAGGTGTCGTTTGCCAAGCTGTAAAAAAGATTGTCGGTGAATGCAGCACTGAACCTACCATTCAGCCTTGTATTCTTGAAGTATCTGATCAGGGGCTTCGTATGGTTGACCGTTCCAAGCCAGAGAGAAATCGCAACGGACCCTGTATCGATTACTTTTACTCGCTGAAGAACGTATCGTTTTGCGCTTTCCATCCTCGAGATCATCGTTACCTGGGTTTCATCACAAAACACCCGACCCTTCAACGATTCGCTTGCCACGTATTTCGAGGACAGGAGTCTACCAGGCCAGTCGCAGAAGCAGTCGGGAGAGCTTTTCAAAGATTTTATCAGAAGTTTATCGAGACAGCGTACCCAATTGAGGACATCTACATAGAATAG
Protein
MADSEFEEFRRVFDRLPQHIKAPTPFYSLVPNVGFEDESPSSKSEGSPEEDRGCVGETDGRSSIPENPESPPPPADAADTQRSPRHQTFTNGERRRRKLPEIPKHKKSSILACGQPASLADELGALTGMLVRPGCHSRPLLVLKCGYLLDEDSSPDSERLQSLGDVDSGHSTAHSPIDTGPKSLSPIPLQQNVSPSSSCSISGMNFAGNMTTVPHSQLEMLEATHRGLHKFNPRHHDEIEVEIGDPIYVQKEAEDLWCEGVNLRTGQQGIFPSAYAVDMDYNDFDPANAKVKRERYLLGYMGSVETLAHKGTGVVCQAVKKIVGECSTEPTIQPCILEVSDQGLRMVDRSKPERNRNGPCIDYFYSLKNVSFCAFHPRDHRYLGFITKHPTLQRFACHVFRGQESTRPVAEAVGRAFQRFYQKFIETAYPIEDIYIE
Summary
Description
The JNK-interacting protein (JIP) group of scaffold proteins selectively mediates JNK signaling by aggregating specific components of the MAPK cascade to form a functional JNK signaling module. May function as a regulator of vesicle transport, through interactions with the JNK-signaling components and motor proteins (By similarity).
Subunit
Forms homo- and heterooligomeric complexes. Binds Hep, a dual specificity protein kinase in the JNK pathway, but not its downstream target bsk. The C-terminal region interacts with the kinesin light chain protein, Klc, and the C-terminal PTY motif of amyloid-beta protein precursor-like protein, Appl.
Similarity
Belongs to the JIP scaffold family.
Keywords
3D-structure
Alternative splicing
Complete proteome
Cytoplasm
Developmental protein
Reference proteome
SH3 domain
Feature
chain JNK-interacting protein 1
splice variant In isoform A.
splice variant In isoform A.
Uniprot
H9ITJ0
A0A2H1VFP4
A0A2A4JMC5
A0A2W1BLU3
A0A212EMR1
A0A194QUK2
+ More
A0A194Q4Z5 A0A0L7LIZ5 Q17KB6 A0A182NKC8 A0A182QM22 A0A0M4EDC6 A0A182KFM5 A0A182YCW8 A0A182IKQ4 A0A182WKJ4 A0A1W4VVN9 A0A182RPF3 Q7Q664 A0A182U4N4 B3M553 A0A084W0Q7 A0A182MUH6 B3NJF3 A0A1Q3FR19 A0A0J9RLW7 Q9W0K0 A0A182I1Q8 A0A182V079 A0A182X775 W5JJI4 A0A1J1I7K8 B4HVN6 A0A182PJJ0 A0A1W4W6Q3 B4IZR1 A0A0Q5U2J0 Q9W0K0-2 A0A0J9RLI6 A0A0P8XTN4 B4PCR5 A0A1I8P0L8 A0A1I8M857 B4KWR3 B4HCQ1 A0A0Q9WIT1 A0A034WPY1 A0A0R3P6Q9 B4HCQ2 B4MM35 A0A3B0KFR2 B4IQZ3 A0A0Q9XBA9 B4HCW6 B4HDF4 A0A0R1DUW6 A0A1I8P0H5 B4LGY8 W8BRE1 B4IQW7 T1PBA5 A0A1I8M853 A0A0L0BVD0 Q29DP0 A0A182FCQ8 A0A034WN93 A0A087ZZD8 D6WHK0 A0A2A3EIS7 B4IR04 W8BAV8 A0A0M9ABH9 A0A1A9Y2A3 B4HD56 A0A1B0GCQ7 A0A1B0C6B2 A0A1B0AD19 B4HCQ3 A0A182SIN2 A0A1W4XE28 A0A195DBW7 A0A151XFP5 A0A195FPZ8 E2AD08 A0A0L7R962 E9IJ96 F4WGX4 A0A195BCD9 A0A158NFE7 B4HDB5 A0A026WKC6 A0A336LRJ2 A0A182GWY6 K7IMD1 E2BF34 A0A0T6B2P9 A0A1B6LGG6 A0A023F351
A0A194Q4Z5 A0A0L7LIZ5 Q17KB6 A0A182NKC8 A0A182QM22 A0A0M4EDC6 A0A182KFM5 A0A182YCW8 A0A182IKQ4 A0A182WKJ4 A0A1W4VVN9 A0A182RPF3 Q7Q664 A0A182U4N4 B3M553 A0A084W0Q7 A0A182MUH6 B3NJF3 A0A1Q3FR19 A0A0J9RLW7 Q9W0K0 A0A182I1Q8 A0A182V079 A0A182X775 W5JJI4 A0A1J1I7K8 B4HVN6 A0A182PJJ0 A0A1W4W6Q3 B4IZR1 A0A0Q5U2J0 Q9W0K0-2 A0A0J9RLI6 A0A0P8XTN4 B4PCR5 A0A1I8P0L8 A0A1I8M857 B4KWR3 B4HCQ1 A0A0Q9WIT1 A0A034WPY1 A0A0R3P6Q9 B4HCQ2 B4MM35 A0A3B0KFR2 B4IQZ3 A0A0Q9XBA9 B4HCW6 B4HDF4 A0A0R1DUW6 A0A1I8P0H5 B4LGY8 W8BRE1 B4IQW7 T1PBA5 A0A1I8M853 A0A0L0BVD0 Q29DP0 A0A182FCQ8 A0A034WN93 A0A087ZZD8 D6WHK0 A0A2A3EIS7 B4IR04 W8BAV8 A0A0M9ABH9 A0A1A9Y2A3 B4HD56 A0A1B0GCQ7 A0A1B0C6B2 A0A1B0AD19 B4HCQ3 A0A182SIN2 A0A1W4XE28 A0A195DBW7 A0A151XFP5 A0A195FPZ8 E2AD08 A0A0L7R962 E9IJ96 F4WGX4 A0A195BCD9 A0A158NFE7 B4HDB5 A0A026WKC6 A0A336LRJ2 A0A182GWY6 K7IMD1 E2BF34 A0A0T6B2P9 A0A1B6LGG6 A0A023F351
Pubmed
19121390
28756777
22118469
26354079
26227816
17510324
+ More
25244985 12364791 14747013 17210077 17994087 24438588 22936249 11912189 10731132 12537572 12537569 20920257 23761445 17550304 25315136 25348373 15632085 24495485 26108605 18362917 19820115 20798317 21282665 21719571 21347285 24508170 30249741 26483478 20075255 25474469
25244985 12364791 14747013 17210077 17994087 24438588 22936249 11912189 10731132 12537572 12537569 20920257 23761445 17550304 25315136 25348373 15632085 24495485 26108605 18362917 19820115 20798317 21282665 21719571 21347285 24508170 30249741 26483478 20075255 25474469
EMBL
BABH01006872
ODYU01002299
SOQ39606.1
NWSH01001075
PCG72734.1
KZ149977
+ More
PZC75888.1 AGBW02013810 OWR42774.1 KQ461108 KPJ09147.1 KQ459463 KPJ00612.1 JTDY01000892 KOB75533.1 CH477226 EAT47131.1 AXCN02000407 CP012525 ALC43584.1 AAAB01008960 EAA11484.4 CH902618 EDV39532.1 ATLV01019151 KE525263 KFB43801.1 AXCM01004843 CH954178 EDV50115.1 GFDL01005026 JAV30019.1 CM002912 KMY96757.1 AF220194 AF231037 AE014296 AY051495 BT001795 APCN01000088 ADMH02001054 ETN64301.1 CVRI01000042 CRK95722.1 CH480817 EDW50001.1 CH916366 EDV95646.1 KQS43051.1 KMY96756.1 KPU78081.1 CM000159 EDW92788.1 CH933809 EDW17510.1 CH479388 EDW29629.1 CH940647 KRF83862.1 GAKP01003144 JAC55808.1 CH379070 KRT08874.1 EDW29630.1 CH963847 EDW73044.2 OUUW01000012 SPP87200.1 CH690070 EDW26662.1 KRG05581.1 CH479486 EDW32958.1 CH480208 EDW26922.1 KRK00719.1 EDW68458.1 GAMC01010744 JAB95811.1 CH689783 EDW39417.1 KA645258 AFP59887.1 JRES01001279 KNC23976.1 EAL30374.1 GAKP01003145 JAC55807.1 KQ971331 EEZ99727.1 KZ288230 PBC31588.1 CH690177 EDW27183.1 GAMC01010743 JAB95812.1 KQ435691 KOX81147.1 CH479720 EDW37585.1 CCAG010002190 JXJN01026526 EDW29631.1 KQ981063 KYN09919.1 KQ982182 KYQ59155.1 KQ981340 KYN42538.1 GL438633 EFN68681.1 KQ414627 KOC67407.1 GL763739 EFZ19364.1 GL888147 EGI66593.1 KQ976514 KYM82226.1 ADTU01014378 ADTU01014379 CH479999 EDW25262.1 KK107199 QOIP01000009 EZA55549.1 RLU18972.1 UFQT01000067 SSX19333.1 JXUM01019983 KQ560561 KXJ81888.1 GL447910 EFN85712.1 LJIG01016061 KRT81731.1 GEBQ01017186 GEBQ01013559 JAT22791.1 JAT26418.1 GBBI01002892 JAC15820.1
PZC75888.1 AGBW02013810 OWR42774.1 KQ461108 KPJ09147.1 KQ459463 KPJ00612.1 JTDY01000892 KOB75533.1 CH477226 EAT47131.1 AXCN02000407 CP012525 ALC43584.1 AAAB01008960 EAA11484.4 CH902618 EDV39532.1 ATLV01019151 KE525263 KFB43801.1 AXCM01004843 CH954178 EDV50115.1 GFDL01005026 JAV30019.1 CM002912 KMY96757.1 AF220194 AF231037 AE014296 AY051495 BT001795 APCN01000088 ADMH02001054 ETN64301.1 CVRI01000042 CRK95722.1 CH480817 EDW50001.1 CH916366 EDV95646.1 KQS43051.1 KMY96756.1 KPU78081.1 CM000159 EDW92788.1 CH933809 EDW17510.1 CH479388 EDW29629.1 CH940647 KRF83862.1 GAKP01003144 JAC55808.1 CH379070 KRT08874.1 EDW29630.1 CH963847 EDW73044.2 OUUW01000012 SPP87200.1 CH690070 EDW26662.1 KRG05581.1 CH479486 EDW32958.1 CH480208 EDW26922.1 KRK00719.1 EDW68458.1 GAMC01010744 JAB95811.1 CH689783 EDW39417.1 KA645258 AFP59887.1 JRES01001279 KNC23976.1 EAL30374.1 GAKP01003145 JAC55807.1 KQ971331 EEZ99727.1 KZ288230 PBC31588.1 CH690177 EDW27183.1 GAMC01010743 JAB95812.1 KQ435691 KOX81147.1 CH479720 EDW37585.1 CCAG010002190 JXJN01026526 EDW29631.1 KQ981063 KYN09919.1 KQ982182 KYQ59155.1 KQ981340 KYN42538.1 GL438633 EFN68681.1 KQ414627 KOC67407.1 GL763739 EFZ19364.1 GL888147 EGI66593.1 KQ976514 KYM82226.1 ADTU01014378 ADTU01014379 CH479999 EDW25262.1 KK107199 QOIP01000009 EZA55549.1 RLU18972.1 UFQT01000067 SSX19333.1 JXUM01019983 KQ560561 KXJ81888.1 GL447910 EFN85712.1 LJIG01016061 KRT81731.1 GEBQ01017186 GEBQ01013559 JAT22791.1 JAT26418.1 GBBI01002892 JAC15820.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000008820 UP000075884 UP000075886 UP000092553 UP000075881 UP000076408 UP000075880 UP000075920 UP000192221 UP000075900 UP000007062 UP000075902 UP000007801 UP000030765 UP000075883 UP000008711 UP000000803 UP000075840 UP000075903 UP000076407 UP000000673 UP000183832 UP000001292 UP000075885 UP000001070 UP000002282 UP000095300 UP000095301 UP000009192 UP000008744 UP000008792 UP000001819 UP000007798 UP000268350 UP000037069 UP000069272 UP000005203 UP000007266 UP000242457 UP000053105 UP000092443 UP000092444 UP000092460 UP000092445 UP000075901 UP000192223 UP000078492 UP000075809 UP000078541 UP000000311 UP000053825 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000069940 UP000249989 UP000002358 UP000008237
UP000008820 UP000075884 UP000075886 UP000092553 UP000075881 UP000076408 UP000075880 UP000075920 UP000192221 UP000075900 UP000007062 UP000075902 UP000007801 UP000030765 UP000075883 UP000008711 UP000000803 UP000075840 UP000075903 UP000076407 UP000000673 UP000183832 UP000001292 UP000075885 UP000001070 UP000002282 UP000095300 UP000095301 UP000009192 UP000008744 UP000008792 UP000001819 UP000007798 UP000268350 UP000037069 UP000069272 UP000005203 UP000007266 UP000242457 UP000053105 UP000092443 UP000092444 UP000092460 UP000092445 UP000075901 UP000192223 UP000078492 UP000075809 UP000078541 UP000000311 UP000053825 UP000007755 UP000078540 UP000005205 UP000053097 UP000279307 UP000069940 UP000249989 UP000002358 UP000008237
Interpro
SUPFAM
SSF50044
SSF50044
Gene 3D
ProteinModelPortal
H9ITJ0
A0A2H1VFP4
A0A2A4JMC5
A0A2W1BLU3
A0A212EMR1
A0A194QUK2
+ More
A0A194Q4Z5 A0A0L7LIZ5 Q17KB6 A0A182NKC8 A0A182QM22 A0A0M4EDC6 A0A182KFM5 A0A182YCW8 A0A182IKQ4 A0A182WKJ4 A0A1W4VVN9 A0A182RPF3 Q7Q664 A0A182U4N4 B3M553 A0A084W0Q7 A0A182MUH6 B3NJF3 A0A1Q3FR19 A0A0J9RLW7 Q9W0K0 A0A182I1Q8 A0A182V079 A0A182X775 W5JJI4 A0A1J1I7K8 B4HVN6 A0A182PJJ0 A0A1W4W6Q3 B4IZR1 A0A0Q5U2J0 Q9W0K0-2 A0A0J9RLI6 A0A0P8XTN4 B4PCR5 A0A1I8P0L8 A0A1I8M857 B4KWR3 B4HCQ1 A0A0Q9WIT1 A0A034WPY1 A0A0R3P6Q9 B4HCQ2 B4MM35 A0A3B0KFR2 B4IQZ3 A0A0Q9XBA9 B4HCW6 B4HDF4 A0A0R1DUW6 A0A1I8P0H5 B4LGY8 W8BRE1 B4IQW7 T1PBA5 A0A1I8M853 A0A0L0BVD0 Q29DP0 A0A182FCQ8 A0A034WN93 A0A087ZZD8 D6WHK0 A0A2A3EIS7 B4IR04 W8BAV8 A0A0M9ABH9 A0A1A9Y2A3 B4HD56 A0A1B0GCQ7 A0A1B0C6B2 A0A1B0AD19 B4HCQ3 A0A182SIN2 A0A1W4XE28 A0A195DBW7 A0A151XFP5 A0A195FPZ8 E2AD08 A0A0L7R962 E9IJ96 F4WGX4 A0A195BCD9 A0A158NFE7 B4HDB5 A0A026WKC6 A0A336LRJ2 A0A182GWY6 K7IMD1 E2BF34 A0A0T6B2P9 A0A1B6LGG6 A0A023F351
A0A194Q4Z5 A0A0L7LIZ5 Q17KB6 A0A182NKC8 A0A182QM22 A0A0M4EDC6 A0A182KFM5 A0A182YCW8 A0A182IKQ4 A0A182WKJ4 A0A1W4VVN9 A0A182RPF3 Q7Q664 A0A182U4N4 B3M553 A0A084W0Q7 A0A182MUH6 B3NJF3 A0A1Q3FR19 A0A0J9RLW7 Q9W0K0 A0A182I1Q8 A0A182V079 A0A182X775 W5JJI4 A0A1J1I7K8 B4HVN6 A0A182PJJ0 A0A1W4W6Q3 B4IZR1 A0A0Q5U2J0 Q9W0K0-2 A0A0J9RLI6 A0A0P8XTN4 B4PCR5 A0A1I8P0L8 A0A1I8M857 B4KWR3 B4HCQ1 A0A0Q9WIT1 A0A034WPY1 A0A0R3P6Q9 B4HCQ2 B4MM35 A0A3B0KFR2 B4IQZ3 A0A0Q9XBA9 B4HCW6 B4HDF4 A0A0R1DUW6 A0A1I8P0H5 B4LGY8 W8BRE1 B4IQW7 T1PBA5 A0A1I8M853 A0A0L0BVD0 Q29DP0 A0A182FCQ8 A0A034WN93 A0A087ZZD8 D6WHK0 A0A2A3EIS7 B4IR04 W8BAV8 A0A0M9ABH9 A0A1A9Y2A3 B4HD56 A0A1B0GCQ7 A0A1B0C6B2 A0A1B0AD19 B4HCQ3 A0A182SIN2 A0A1W4XE28 A0A195DBW7 A0A151XFP5 A0A195FPZ8 E2AD08 A0A0L7R962 E9IJ96 F4WGX4 A0A195BCD9 A0A158NFE7 B4HDB5 A0A026WKC6 A0A336LRJ2 A0A182GWY6 K7IMD1 E2BF34 A0A0T6B2P9 A0A1B6LGG6 A0A023F351
PDB
2FPE
E-value=2.46276e-13,
Score=183
Ontologies
GO
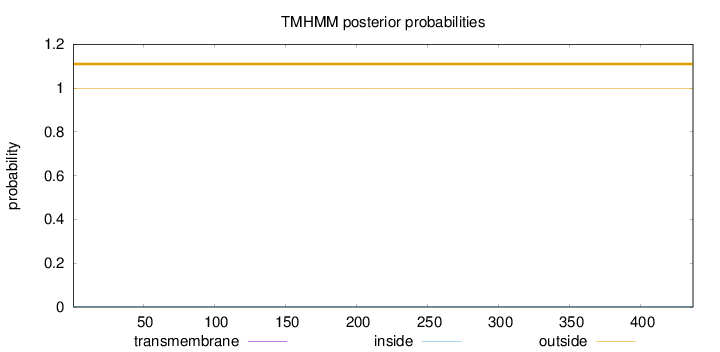
Topology
Subcellular location
Cytoplasm
Length:
437
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000460000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00213
outside
1 - 437
Population Genetic Test Statistics
Pi
5.873898
Theta
13.098943
Tajima's D
-1.55012
CLR
0
CSRT
0.0527973601319934
Interpretation
Uncertain
