Gene
KWMTBOMO00224
Pre Gene Modal
BGIBMGA000640
Annotation
cytochrome_P450_Cyp305B1_precursor_[Bombyx_mori]
Full name
Probable cytochrome P450 305a1
Alternative Name
CYPCCCVA1
Location in the cell
PlasmaMembrane Reliability : 3.178
Sequence
CDS
ATGCTGCCAGTGCTTGTTTGCATAATAGTTATCGTATTAATCTGTTGTAATGTCATCAGAAGTGTAATCAAACCCGAGAAATTTCCTCCAGGTCCGATATGGTATCCATTTTTTGGCAGCAGCTCAATTGTTCAACAAATGACAAGCAAACACGGGTCGCAGTGGAAAGCGTTACTGGAACTTTCGAAACAGTGGTCCACTCAGGTTCTCGGCTTAAAACTAGGAAGAGAATTAGTGGTCGTCGTCTACGGGGAGAAGAATGTTCGTCAAGTGTTCTCGGAATCCGAGTTCGACGGTCGACCGAACTCTTTCTTCTATAAGTTGAGATGTCTCGGAAAACGACTCGGCGTCACCTTTGTTGATGGTCCTCTTTGGCGAGAACACAGACAGTTCACTGTAAAACATTTAAAGAATGTTGGTTTTGGTAAGACATCGATGGAGCTGGAAATCCAAAACGAATTGAAGTTGTTACGTGAATACATAAATGATAATAAACATAAGCCAATAAAAGTTGATAGTATGTTTTCATCGGCCGTAATGAATGTTTTATGGAAGTATGTAGCAGGCGAGAGAATCAGAGAAGACAAATTAGAACGTTTACTTGAACTATTCTATTTGAGATCGAAGGCGTTCACTTTGACTGGTGGCTTGCTAAGCCAAATACCTTGGTGCAGGTTCATTATACCCGGCCTAAGTGGGTACAAACTAATAGTTGACTTGAACCAGCAGATATCTGAAATTATTGAGGAAGCCATAAAAAAGCATTTAAACAAAGAAGTGCAACAGAACGATTTTATATATTCGTTCTTAGATGAAATGAACGAAGAGAACAAGGCATCATTTACGTACGATCAGCTGAAAACCGTGTGCCTTGATCTAATAATAGCCGGATCCCAAACAACAGGGAACGCTGTTAAGTTTGCTCTTCTATCAGTTTTGCGAAACAAAAACATTCAAGAAAAAATCTTCAACGAAATAGAAAACACCATTGGCGATTCTATGCCTTGCTGGGCTGACAGTAGCAAACTCGTATATACATCGGCATTTCTGCTGGAAGTAATGAGAATTCACACAATAGCGCCCCTGGCTGGTCCTCGAAGAGTTTTACAGGACACAGTCATCGACGGCTACGTTATACCTAAAGAGACGACGGTTCTGATATCCTTGGCCGATATTCACTTGGATCCGAATCTGTGGCCGGATCCTCACGAAATAAAGCCGGAGAGGTTTATCGACGAAAAAGGATTATCGAAAAGTAATGAACACATCTATCCTTTTGGATCAGGTCGGAGACGTTGCCCAGGCGATTCGCTGGCCAGGTCTTTCGTCTTTATCATATTTGTTGGGATCCTGCAGAAGTACAGAATAGACTGTGTCAATGGAGTATTGCCGTCTAACGAAGCTGACATAGGCTTGCTTGCGGCTCCAAAACCTTTTGTCGCTAATTTCGTATCAAGAGAGTAA
Protein
MLPVLVCIIVIVLICCNVIRSVIKPEKFPPGPIWYPFFGSSSIVQQMTSKHGSQWKALLELSKQWSTQVLGLKLGRELVVVVYGEKNVRQVFSESEFDGRPNSFFYKLRCLGKRLGVTFVDGPLWREHRQFTVKHLKNVGFGKTSMELEIQNELKLLREYINDNKHKPIKVDSMFSSAVMNVLWKYVAGERIREDKLERLLELFYLRSKAFTLTGGLLSQIPWCRFIIPGLSGYKLIVDLNQQISEIIEEAIKKHLNKEVQQNDFIYSFLDEMNEENKASFTYDQLKTVCLDLIIAGSQTTGNAVKFALLSVLRNKNIQEKIFNEIENTIGDSMPCWADSSKLVYTSAFLLEVMRIHTIAPLAGPRRVLQDTVIDGYVIPKETTVLISLADIHLDPNLWPDPHEIKPERFIDEKGLSKSNEHIYPFGSGRRRCPGDSLARSFVFIIFVGILQKYRIDCVNGVLPSNEADIGLLAAPKPFVANFVSRE
Summary
Description
May be involved in the metabolism of insect hormones and in the breakdown of synthetic insecticides.
Cofactor
heme
Similarity
Belongs to the cytochrome P450 family.
Keywords
Complete proteome
Endoplasmic reticulum
Heme
Iron
Membrane
Metal-binding
Microsome
Monooxygenase
Oxidoreductase
Reference proteome
Feature
chain Probable cytochrome P450 305a1
Uniprot
L0N6J8
Q2L9Y9
Q9NDL3
H9ITR0
A0A0C5C541
A0A248QEI5
+ More
A0A1V0D9F4 T1WK13 A0A2W1BLF5 A0A068EU19 A0A194QB17 A0A194QZY0 A0A212EMV4 X5D4F5 A0A286MXN2 A0A212EMN8 Q17JA9 A0A1W5KJU1 Q17JB0 A0A336N4L8 A0A336K893 A0A1J1IIJ5 A0A1B6F8W0 A0A1B0FC64 A0A1Q3G500 A0A182PZ34 A0A1B6HAC5 A0A0K1YWH2 A0A068F800 A0A1Y1JZV7 A0A0A1WHE4 A0A0M3QWH8 A0A182LX08 A0A1J1I412 A0A182QNR3 A0A182RFA6 A0A3F2Z158 B4L0X7 A0A2J7QMN4 A0A1W4WN12 A0A1A9WS92 A0A0L0C6F8 D6WFY5 A0A084W481 B3NIJ0 A0A3F2YW28 A0A248ADT7 A0A1B0C6V2 B4LBM9 A0A0K8VYF8 A0A290GNU1 A0A084W482 A0A182KC89 A0A1A9Z3L0 A0A182Y1B2 A0A182PHN4 W5JLT1 A0A034VQK2 A0A1Q1NL12 A0A1A9XBI4 A0A0K0LB87 A0A1A9UXW5 A0A125S6P6 A0A182UIL2 A0A182XUJ9 Q7Q6U9 A0A182HWR4 Q5TRI3 A0A182FZX1 K4JN85 A0A1I8MUR4 B4QQY9 Q7QLM0 W5JL39 A0A182FZX0 B4IXQ0 B3M8U6 Q9VW43 A0A182XUK0 A0A182UV89 B4IIQ9 A0A0K0LC82 A0A182VJM2 A0A1B6BXF9 A0A0K8SBH9 A0A067QG21 A0A0A9XER8 J9K9I4 A0A1I8NQB8 A0A182NGX6 A0A182Y1B3 B4H921 B4IUD3 Q2M000 A0A1B0DPF7 B4PFX8 Q7PIP7 A0A1W4VUW0 B0X5V2 N6U564
A0A1V0D9F4 T1WK13 A0A2W1BLF5 A0A068EU19 A0A194QB17 A0A194QZY0 A0A212EMV4 X5D4F5 A0A286MXN2 A0A212EMN8 Q17JA9 A0A1W5KJU1 Q17JB0 A0A336N4L8 A0A336K893 A0A1J1IIJ5 A0A1B6F8W0 A0A1B0FC64 A0A1Q3G500 A0A182PZ34 A0A1B6HAC5 A0A0K1YWH2 A0A068F800 A0A1Y1JZV7 A0A0A1WHE4 A0A0M3QWH8 A0A182LX08 A0A1J1I412 A0A182QNR3 A0A182RFA6 A0A3F2Z158 B4L0X7 A0A2J7QMN4 A0A1W4WN12 A0A1A9WS92 A0A0L0C6F8 D6WFY5 A0A084W481 B3NIJ0 A0A3F2YW28 A0A248ADT7 A0A1B0C6V2 B4LBM9 A0A0K8VYF8 A0A290GNU1 A0A084W482 A0A182KC89 A0A1A9Z3L0 A0A182Y1B2 A0A182PHN4 W5JLT1 A0A034VQK2 A0A1Q1NL12 A0A1A9XBI4 A0A0K0LB87 A0A1A9UXW5 A0A125S6P6 A0A182UIL2 A0A182XUJ9 Q7Q6U9 A0A182HWR4 Q5TRI3 A0A182FZX1 K4JN85 A0A1I8MUR4 B4QQY9 Q7QLM0 W5JL39 A0A182FZX0 B4IXQ0 B3M8U6 Q9VW43 A0A182XUK0 A0A182UV89 B4IIQ9 A0A0K0LC82 A0A182VJM2 A0A1B6BXF9 A0A0K8SBH9 A0A067QG21 A0A0A9XER8 J9K9I4 A0A1I8NQB8 A0A182NGX6 A0A182Y1B3 B4H921 B4IUD3 Q2M000 A0A1B0DPF7 B4PFX8 Q7PIP7 A0A1W4VUW0 B0X5V2 N6U564
EC Number
1.14.-.-
Pubmed
19121390
28881445
22984814
28756777
26354079
22118469
+ More
24361403 19954531 25299618 17510324 25880816 28004739 25830018 17994087 26108605 18362917 19820115 24438588 25244985 20920257 23761445 25348373 28092127 12364791 14747013 17210077 24324548 25315136 22936249 9087549 10731132 12537572 12537569 24845553 25401762 17550304 15632085 23537049
24361403 19954531 25299618 17510324 25880816 28004739 25830018 17994087 26108605 18362917 19820115 24438588 25244985 20920257 23761445 25348373 28092127 12364791 14747013 17210077 24324548 25315136 22936249 9087549 10731132 12537572 12537569 24845553 25401762 17550304 15632085 23537049
EMBL
AK289325
BAM73852.1
DQ342362
ABC69163.1
DQ417659
AB044900
+ More
ABD78849.1 BAA96811.1 BABH01006864 BABH01006865 KP001120 AJN91165.1 KX443454 ASO98029.1 KY212065 ARA91629.1 JQ979047 AGU36301.1 KZ149977 PZC75879.1 KM016702 AID54854.1 KQ459463 KPJ00616.1 KQ461108 KPJ09151.1 AGBW02013811 OWR42771.1 KF701126 AHW57296.1 MF684350 ASX93984.1 OWR42770.1 CH477233 EAT46796.1 KU941587 AMR44690.1 EAT46795.1 UFQS01002401 UFQT01002401 SSX13959.1 SSX33378.1 UFQS01000067 UFQT01000067 SSW98983.1 SSX19365.1 CVRI01000054 CRL00053.1 GECZ01023087 JAS46682.1 CCAG010016073 GFDL01000171 JAV34874.1 GECU01036119 JAS71587.1 KP859378 AKZ17688.1 KJ702243 AID61397.1 GEZM01098710 JAV53791.1 GBXI01016362 JAC97929.1 CP012525 ALC44173.1 AXCM01013784 CVRI01000040 CRK95079.1 AXCN02000360 CH933809 EDW19227.1 NEVH01013204 PNF29828.1 JRES01000836 KNC27860.1 KQ971328 EFA01265.1 ATLV01020285 KE525297 KFB45025.1 CH954178 EDV52415.1 KX708494 ARG41841.1 JXJN01027023 JXJN01027024 CH940647 EDW70839.1 GDHF01034073 GDHF01008366 JAI18241.1 JAI43948.1 KX945358 ATB55046.1 KFB45026.1 ADMH02000773 ETN65091.1 GAKP01014238 JAC44714.1 KX660731 AQM57070.1 KM217012 AIW79975.1 KT601138 AME15807.1 AAAB01008960 EAA11827.3 APCN01001532 EAL39881.2 JX876511 AFU86441.1 CM000363 CM002912 EDX11127.1 KMZ00615.1 AAAB01006489 EAA02774.4 ETN65092.1 CH916366 EDV97512.1 CH902618 EDV41097.1 AE014296 AY051626 CH480844 EDW49830.1 KM217011 AIW79974.1 GEDC01031608 JAS05690.1 GBRD01015197 GBRD01015196 GBRD01015194 GBRD01015193 JAG50633.1 KK853574 KDR06612.1 GBHO01026291 GBHO01026290 GBHO01026288 GBHO01026286 JAG17313.1 JAG17314.1 JAG17316.1 JAG17318.1 ABLF02021786 CH479226 EDW35236.1 CH891868 EDW99996.1 CH379069 EAL31137.1 AJVK01008091 CM000159 EDW95275.1 EAA44065.3 DS232396 EDS41083.1 APGK01039300 APGK01039301 KB740969 ENN76740.1
ABD78849.1 BAA96811.1 BABH01006864 BABH01006865 KP001120 AJN91165.1 KX443454 ASO98029.1 KY212065 ARA91629.1 JQ979047 AGU36301.1 KZ149977 PZC75879.1 KM016702 AID54854.1 KQ459463 KPJ00616.1 KQ461108 KPJ09151.1 AGBW02013811 OWR42771.1 KF701126 AHW57296.1 MF684350 ASX93984.1 OWR42770.1 CH477233 EAT46796.1 KU941587 AMR44690.1 EAT46795.1 UFQS01002401 UFQT01002401 SSX13959.1 SSX33378.1 UFQS01000067 UFQT01000067 SSW98983.1 SSX19365.1 CVRI01000054 CRL00053.1 GECZ01023087 JAS46682.1 CCAG010016073 GFDL01000171 JAV34874.1 GECU01036119 JAS71587.1 KP859378 AKZ17688.1 KJ702243 AID61397.1 GEZM01098710 JAV53791.1 GBXI01016362 JAC97929.1 CP012525 ALC44173.1 AXCM01013784 CVRI01000040 CRK95079.1 AXCN02000360 CH933809 EDW19227.1 NEVH01013204 PNF29828.1 JRES01000836 KNC27860.1 KQ971328 EFA01265.1 ATLV01020285 KE525297 KFB45025.1 CH954178 EDV52415.1 KX708494 ARG41841.1 JXJN01027023 JXJN01027024 CH940647 EDW70839.1 GDHF01034073 GDHF01008366 JAI18241.1 JAI43948.1 KX945358 ATB55046.1 KFB45026.1 ADMH02000773 ETN65091.1 GAKP01014238 JAC44714.1 KX660731 AQM57070.1 KM217012 AIW79975.1 KT601138 AME15807.1 AAAB01008960 EAA11827.3 APCN01001532 EAL39881.2 JX876511 AFU86441.1 CM000363 CM002912 EDX11127.1 KMZ00615.1 AAAB01006489 EAA02774.4 ETN65092.1 CH916366 EDV97512.1 CH902618 EDV41097.1 AE014296 AY051626 CH480844 EDW49830.1 KM217011 AIW79974.1 GEDC01031608 JAS05690.1 GBRD01015197 GBRD01015196 GBRD01015194 GBRD01015193 JAG50633.1 KK853574 KDR06612.1 GBHO01026291 GBHO01026290 GBHO01026288 GBHO01026286 JAG17313.1 JAG17314.1 JAG17316.1 JAG17318.1 ABLF02021786 CH479226 EDW35236.1 CH891868 EDW99996.1 CH379069 EAL31137.1 AJVK01008091 CM000159 EDW95275.1 EAA44065.3 DS232396 EDS41083.1 APGK01039300 APGK01039301 KB740969 ENN76740.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000008820
UP000183832
+ More
UP000092444 UP000075885 UP000092553 UP000075883 UP000075886 UP000075900 UP000075920 UP000009192 UP000235965 UP000192223 UP000091820 UP000037069 UP000007266 UP000030765 UP000008711 UP000075884 UP000092460 UP000008792 UP000075881 UP000092445 UP000076408 UP000000673 UP000092443 UP000078200 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000095301 UP000000304 UP000001070 UP000007801 UP000000803 UP000075903 UP000001292 UP000027135 UP000007819 UP000095300 UP000008744 UP000002282 UP000001819 UP000092462 UP000192221 UP000002320 UP000019118
UP000092444 UP000075885 UP000092553 UP000075883 UP000075886 UP000075900 UP000075920 UP000009192 UP000235965 UP000192223 UP000091820 UP000037069 UP000007266 UP000030765 UP000008711 UP000075884 UP000092460 UP000008792 UP000075881 UP000092445 UP000076408 UP000000673 UP000092443 UP000078200 UP000075902 UP000076407 UP000007062 UP000075840 UP000069272 UP000095301 UP000000304 UP000001070 UP000007801 UP000000803 UP000075903 UP000001292 UP000027135 UP000007819 UP000095300 UP000008744 UP000002282 UP000001819 UP000092462 UP000192221 UP000002320 UP000019118
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
L0N6J8
Q2L9Y9
Q9NDL3
H9ITR0
A0A0C5C541
A0A248QEI5
+ More
A0A1V0D9F4 T1WK13 A0A2W1BLF5 A0A068EU19 A0A194QB17 A0A194QZY0 A0A212EMV4 X5D4F5 A0A286MXN2 A0A212EMN8 Q17JA9 A0A1W5KJU1 Q17JB0 A0A336N4L8 A0A336K893 A0A1J1IIJ5 A0A1B6F8W0 A0A1B0FC64 A0A1Q3G500 A0A182PZ34 A0A1B6HAC5 A0A0K1YWH2 A0A068F800 A0A1Y1JZV7 A0A0A1WHE4 A0A0M3QWH8 A0A182LX08 A0A1J1I412 A0A182QNR3 A0A182RFA6 A0A3F2Z158 B4L0X7 A0A2J7QMN4 A0A1W4WN12 A0A1A9WS92 A0A0L0C6F8 D6WFY5 A0A084W481 B3NIJ0 A0A3F2YW28 A0A248ADT7 A0A1B0C6V2 B4LBM9 A0A0K8VYF8 A0A290GNU1 A0A084W482 A0A182KC89 A0A1A9Z3L0 A0A182Y1B2 A0A182PHN4 W5JLT1 A0A034VQK2 A0A1Q1NL12 A0A1A9XBI4 A0A0K0LB87 A0A1A9UXW5 A0A125S6P6 A0A182UIL2 A0A182XUJ9 Q7Q6U9 A0A182HWR4 Q5TRI3 A0A182FZX1 K4JN85 A0A1I8MUR4 B4QQY9 Q7QLM0 W5JL39 A0A182FZX0 B4IXQ0 B3M8U6 Q9VW43 A0A182XUK0 A0A182UV89 B4IIQ9 A0A0K0LC82 A0A182VJM2 A0A1B6BXF9 A0A0K8SBH9 A0A067QG21 A0A0A9XER8 J9K9I4 A0A1I8NQB8 A0A182NGX6 A0A182Y1B3 B4H921 B4IUD3 Q2M000 A0A1B0DPF7 B4PFX8 Q7PIP7 A0A1W4VUW0 B0X5V2 N6U564
A0A1V0D9F4 T1WK13 A0A2W1BLF5 A0A068EU19 A0A194QB17 A0A194QZY0 A0A212EMV4 X5D4F5 A0A286MXN2 A0A212EMN8 Q17JA9 A0A1W5KJU1 Q17JB0 A0A336N4L8 A0A336K893 A0A1J1IIJ5 A0A1B6F8W0 A0A1B0FC64 A0A1Q3G500 A0A182PZ34 A0A1B6HAC5 A0A0K1YWH2 A0A068F800 A0A1Y1JZV7 A0A0A1WHE4 A0A0M3QWH8 A0A182LX08 A0A1J1I412 A0A182QNR3 A0A182RFA6 A0A3F2Z158 B4L0X7 A0A2J7QMN4 A0A1W4WN12 A0A1A9WS92 A0A0L0C6F8 D6WFY5 A0A084W481 B3NIJ0 A0A3F2YW28 A0A248ADT7 A0A1B0C6V2 B4LBM9 A0A0K8VYF8 A0A290GNU1 A0A084W482 A0A182KC89 A0A1A9Z3L0 A0A182Y1B2 A0A182PHN4 W5JLT1 A0A034VQK2 A0A1Q1NL12 A0A1A9XBI4 A0A0K0LB87 A0A1A9UXW5 A0A125S6P6 A0A182UIL2 A0A182XUJ9 Q7Q6U9 A0A182HWR4 Q5TRI3 A0A182FZX1 K4JN85 A0A1I8MUR4 B4QQY9 Q7QLM0 W5JL39 A0A182FZX0 B4IXQ0 B3M8U6 Q9VW43 A0A182XUK0 A0A182UV89 B4IIQ9 A0A0K0LC82 A0A182VJM2 A0A1B6BXF9 A0A0K8SBH9 A0A067QG21 A0A0A9XER8 J9K9I4 A0A1I8NQB8 A0A182NGX6 A0A182Y1B3 B4H921 B4IUD3 Q2M000 A0A1B0DPF7 B4PFX8 Q7PIP7 A0A1W4VUW0 B0X5V2 N6U564
PDB
3DL9
E-value=3.43014e-48,
Score=484
Ontologies
GO
Topology
Subcellular location
Endoplasmic reticulum membrane
Microsome membrane
Microsome membrane
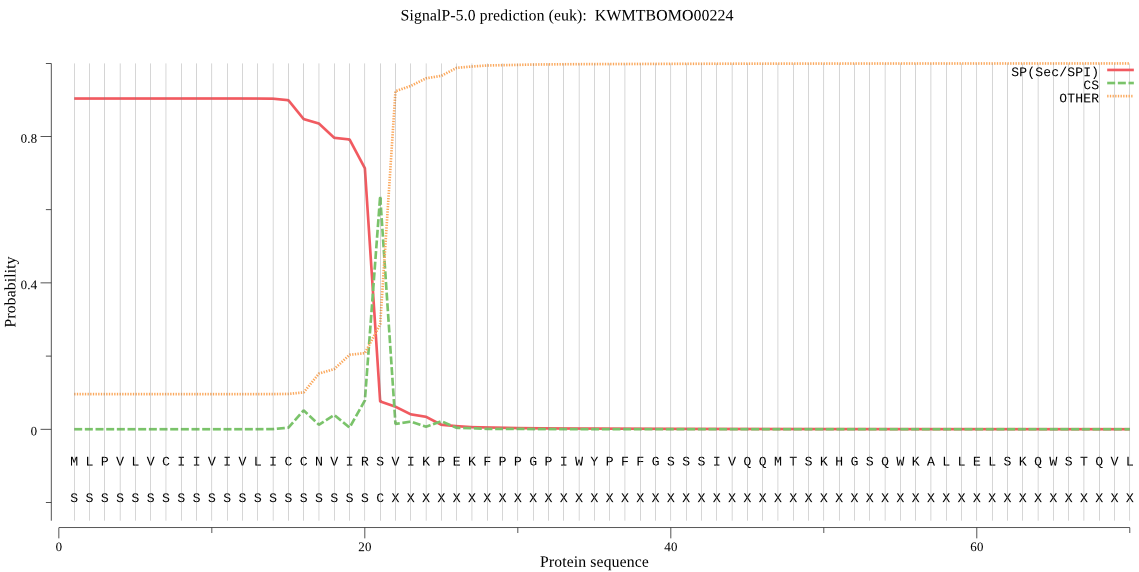
SignalP
Position: 1 - 21,
Likelihood: 0.903595
Length:
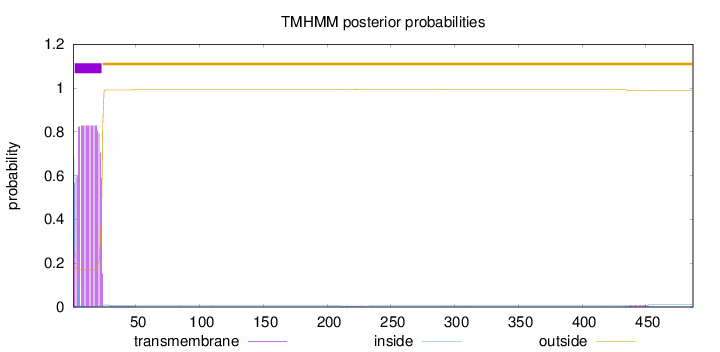
487
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
17.4823199999999
Exp number, first 60 AAs:
17.29076
Total prob of N-in:
0.82041
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 23
outside
24 - 487
Population Genetic Test Statistics
Pi
22.677114
Theta
24.930991
Tajima's D
-0.767903
CLR
0.053699
CSRT
0.187890605469727
Interpretation
Uncertain
