Gene
KWMTBOMO00221
Pre Gene Modal
BGIBMGA000639
Annotation
PREDICTED:_homeobox_protein_caupolican_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 3.999
Sequence
CDS
ATGAGCGCGGGAGCGGGTAGCGCGGCTCGGCCTAGCTCGCCCGCCGCCGCGCCTCGCTGCTGTGACACCGGACGCCCCATATTCCACGACCCGATCACGGGCCAGACGGTCTGCTCCTGTCAATACGAGTTCCTAAACTACCAGAGGCTAGCTTCGGGCATGCCTATGTCTGTGTACAGTGCCCCCTACTCCGACGCGGCTGCTGCAACGGCAGCCGGCATGGCAGCATACTTTCCTGCCCTAGCTGCCGACCAACCTCCATTTTACACAAACGCGGCAGCGGGAATCGAGCTGAAAGAAAATTTAGCAGCCAGCGCAGCGAGCTGGCCTTATCCTGCAGTCTATCATCCTTACGATGCGGCTTTTGCTGGATATCCCTTCAACGGGTACGGGATGGACCTAAACGGAGCGCGAAGGAAAAATGCTACGCGAGAAACGACTAGCACGCTCAAAGCATGGCTCAACGAGCACAAGAAGAATCCTTATCCAACGAAAGGGGAGAAGATTATGCTTGCTATCATTACCAAAATGACGCTTACGCAAGTTTCTACTTGGTTTGCGAATGCGCGTCGTCGACTGAAGAAAGAGAACAAGATGACGTGGGAACCAAGAAACAGGGTTGACGATGATGACAATAACAATGACGACGATGACCACAAAAGTAATGACGGCAAAGACGCTTTAGATGGTAAGGACTCCGGCACAGGTTCGAGTGAAGACGGAGACAGACCTCAGCAGAGGTTGGATCTTTTGGGACAACGAACAGAATCAGAGTGGTCAGAATCACGAGCGGACAGCGGACCTGAGTCACCTGAGCCGTATGAGAGACCGATACATCCGGCGTACCAACATTTACCTTCCCGCGCACCACCTGGCAGCACCCCTGCATCGGCCAAGCCCAGAATATGGTCGCTAGCGGATATGGCGAGCAAAGACGGTGAGTCGCCTGCGCCTCCGTCATCGGCTTCCGCTTTCTACCAGTCAGCTGCAGCGGCTGCAGCGGCTGCGCGATTAGCACATCCTTACGGAAGGCCCGAACTATATCGAGGGCTATATCCTCCAACTCACCCCGCTGACGTGGCTCTGTTGGAATATTCTCGGTCTTTAGCTCTCGCTAATCCAGCACCTGCACCGCCGGCTCCTTCACCGTCGTCTTCATCGACATCGTCAGTGGCCGAGCCGCCACCACCGCCCCGTGCCTGA
Protein
MSAGAGSAARPSSPAAAPRCCDTGRPIFHDPITGQTVCSCQYEFLNYQRLASGMPMSVYSAPYSDAAAATAAGMAAYFPALAADQPPFYTNAAAGIELKENLAASAASWPYPAVYHPYDAAFAGYPFNGYGMDLNGARRKNATRETTSTLKAWLNEHKKNPYPTKGEKIMLAIITKMTLTQVSTWFANARRRLKKENKMTWEPRNRVDDDDNNNDDDDHKSNDGKDALDGKDSGTGSSEDGDRPQQRLDLLGQRTESEWSESRADSGPESPEPYERPIHPAYQHLPSRAPPGSTPASAKPRIWSLADMASKDGESPAPPSSASAFYQSAAAAAAAARLAHPYGRPELYRGLYPPTHPADVALLEYSRSLALANPAPAPPAPSPSSSSTSSVAEPPPPPRA
Summary
Uniprot
A0A2W1BNZ6
A0A2A4JJ46
A0A2A4JKQ6
A0A139WL71
U4UQ21
N6U4R4
+ More
A0A1Y1LTB9 A0A1B0EWI7 A0A1S4FGP2 Q171W7 B0VZ28 A0A182GVJ9 A0A2M4CRT0 A0A2M4CRZ8 A0A154P8Q5 A0A084WDT6 A0A151J012 A0A1W4WZT5 A0A1W4WYS6 A0A1W4WP17 A0A195FNQ3 F4WMP6 A0A158N8X2 A0A1Y9H1X5 A0A2C9H895 A0A3F2Z0C0 Q7Q7U9 A0A2C9GPV8 A0A195B4P8 A0A1B6E4I8 A0A088AU13 A0A0C9RPT6 A0A0C9QIC1 A0A0L7QZW5 A0A067R8U0 A0A3L8DDD6 A0A2S2QBZ6 A0A026W5Y9 A0A3B0KMM3 K7IW51 A0A1W4UH91 B4NDK3 A0A1J1IJB5 A0A1B6GLA3 B4GRH6 A0A1B6M728 Q2M0K9 B3M9P9 B4HG50 B4QRQ3 O01667 Q9VU01 A0A0J9RVI5 B4PGT5 M9PI45 B3NCT4 A0A0M4EZX6 A0A1S4EBL4 B4IWZ1 B4LGH0 A0A0A9W3F2 W8B7E1 B4L0P7 A0A0A1WF40 A0A0K8UJ47 T1GN44 A0A1I8M686 A0A1I8M684 A0A336MWQ2 A0A336M4F8 A0A1B0FA50 A0A0L0C998 A0A0A9VZE3 A0A034VZD6 A0A2R7VVJ0
A0A1Y1LTB9 A0A1B0EWI7 A0A1S4FGP2 Q171W7 B0VZ28 A0A182GVJ9 A0A2M4CRT0 A0A2M4CRZ8 A0A154P8Q5 A0A084WDT6 A0A151J012 A0A1W4WZT5 A0A1W4WYS6 A0A1W4WP17 A0A195FNQ3 F4WMP6 A0A158N8X2 A0A1Y9H1X5 A0A2C9H895 A0A3F2Z0C0 Q7Q7U9 A0A2C9GPV8 A0A195B4P8 A0A1B6E4I8 A0A088AU13 A0A0C9RPT6 A0A0C9QIC1 A0A0L7QZW5 A0A067R8U0 A0A3L8DDD6 A0A2S2QBZ6 A0A026W5Y9 A0A3B0KMM3 K7IW51 A0A1W4UH91 B4NDK3 A0A1J1IJB5 A0A1B6GLA3 B4GRH6 A0A1B6M728 Q2M0K9 B3M9P9 B4HG50 B4QRQ3 O01667 Q9VU01 A0A0J9RVI5 B4PGT5 M9PI45 B3NCT4 A0A0M4EZX6 A0A1S4EBL4 B4IWZ1 B4LGH0 A0A0A9W3F2 W8B7E1 B4L0P7 A0A0A1WF40 A0A0K8UJ47 T1GN44 A0A1I8M686 A0A1I8M684 A0A336MWQ2 A0A336M4F8 A0A1B0FA50 A0A0L0C998 A0A0A9VZE3 A0A034VZD6 A0A2R7VVJ0
Pubmed
28756777
18362917
19820115
23537049
28004739
17510324
+ More
26483478 24438588 21719571 21347285 12364791 14747013 17210077 24845553 30249741 24508170 20075255 17994087 15632085 9136934 9477320 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 25401762 24495485 25830018 25315136 26108605 25348373
26483478 24438588 21719571 21347285 12364791 14747013 17210077 24845553 30249741 24508170 20075255 17994087 15632085 9136934 9477320 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 25401762 24495485 25830018 25315136 26108605 25348373
EMBL
KZ149973
PZC75991.1
NWSH01001244
PCG71991.1
PCG71990.1
KQ971321
+ More
KYB28759.1 KB632398 ERL94608.1 APGK01042656 APGK01042657 APGK01042658 APGK01042659 APGK01042660 KB741007 ENN75616.1 GEZM01052924 JAV74237.1 AJWK01005725 AJWK01005726 AJWK01005727 AJWK01005728 CH477444 EAT40788.1 DS231813 EDS25750.1 JXUM01018967 KQ560529 KXJ81971.1 GGFL01003868 MBW68046.1 GGFL01003867 MBW68045.1 KQ434846 KZC08299.1 ATLV01023063 KE525340 KFB48380.1 KQ980658 KYN14747.1 KQ981382 KYN42180.1 GL888218 EGI64523.1 ADTU01008893 ADTU01008894 ADTU01008895 AAAB01008948 EAA10468.4 APCN01004682 KQ976618 KYM79242.1 GEDC01004474 JAS32824.1 GBYB01010495 JAG80262.1 GBYB01014503 JAG84270.1 KQ414676 KOC64096.1 KK852664 KDR19021.1 QOIP01000010 RLU18163.1 GGMS01006076 MBY75279.1 KK107455 EZA50454.1 OUUW01000009 SPP85058.1 CH964239 EDW81825.2 CVRI01000054 CRL00333.1 GECZ01006589 JAS63180.1 CH479188 EDW40361.1 GEBQ01008245 JAT31732.1 CH379069 EAL30921.2 CH902618 EDV39055.2 CH480815 EDW41296.1 CM000363 EDX10267.1 U95021 AF004710 AAB53640.1 AAC23943.1 AE014296 AAF49894.1 AAN11850.1 CM002912 KMY99269.1 CM000159 EDW94324.2 AGB94471.1 CH954178 EDV51590.2 CP012525 ALC44350.1 CH916366 EDV97392.1 CH940647 EDW70499.2 GBHO01042626 JAG00978.1 GAMC01013607 JAB92948.1 CH933809 EDW18124.2 GBXI01017197 JAC97094.1 GDHF01025728 JAI26586.1 CAQQ02395044 UFQT01003166 SSX34676.1 UFQT01000549 SSX25162.1 CCAG010009438 JRES01000715 KNC28998.1 GBHO01042625 JAG00979.1 GAKP01011742 JAC47210.1 KK854083 PTY10911.1
KYB28759.1 KB632398 ERL94608.1 APGK01042656 APGK01042657 APGK01042658 APGK01042659 APGK01042660 KB741007 ENN75616.1 GEZM01052924 JAV74237.1 AJWK01005725 AJWK01005726 AJWK01005727 AJWK01005728 CH477444 EAT40788.1 DS231813 EDS25750.1 JXUM01018967 KQ560529 KXJ81971.1 GGFL01003868 MBW68046.1 GGFL01003867 MBW68045.1 KQ434846 KZC08299.1 ATLV01023063 KE525340 KFB48380.1 KQ980658 KYN14747.1 KQ981382 KYN42180.1 GL888218 EGI64523.1 ADTU01008893 ADTU01008894 ADTU01008895 AAAB01008948 EAA10468.4 APCN01004682 KQ976618 KYM79242.1 GEDC01004474 JAS32824.1 GBYB01010495 JAG80262.1 GBYB01014503 JAG84270.1 KQ414676 KOC64096.1 KK852664 KDR19021.1 QOIP01000010 RLU18163.1 GGMS01006076 MBY75279.1 KK107455 EZA50454.1 OUUW01000009 SPP85058.1 CH964239 EDW81825.2 CVRI01000054 CRL00333.1 GECZ01006589 JAS63180.1 CH479188 EDW40361.1 GEBQ01008245 JAT31732.1 CH379069 EAL30921.2 CH902618 EDV39055.2 CH480815 EDW41296.1 CM000363 EDX10267.1 U95021 AF004710 AAB53640.1 AAC23943.1 AE014296 AAF49894.1 AAN11850.1 CM002912 KMY99269.1 CM000159 EDW94324.2 AGB94471.1 CH954178 EDV51590.2 CP012525 ALC44350.1 CH916366 EDV97392.1 CH940647 EDW70499.2 GBHO01042626 JAG00978.1 GAMC01013607 JAB92948.1 CH933809 EDW18124.2 GBXI01017197 JAC97094.1 GDHF01025728 JAI26586.1 CAQQ02395044 UFQT01003166 SSX34676.1 UFQT01000549 SSX25162.1 CCAG010009438 JRES01000715 KNC28998.1 GBHO01042625 JAG00979.1 GAKP01011742 JAC47210.1 KK854083 PTY10911.1
Proteomes
UP000218220
UP000007266
UP000030742
UP000019118
UP000092461
UP000008820
+ More
UP000002320 UP000069940 UP000249989 UP000076502 UP000030765 UP000078492 UP000192223 UP000078541 UP000007755 UP000005205 UP000075884 UP000076407 UP000075903 UP000007062 UP000075840 UP000078540 UP000005203 UP000053825 UP000027135 UP000279307 UP000053097 UP000268350 UP000002358 UP000192221 UP000007798 UP000183832 UP000008744 UP000001819 UP000007801 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000092553 UP000079169 UP000001070 UP000008792 UP000009192 UP000015102 UP000095301 UP000092444 UP000037069
UP000002320 UP000069940 UP000249989 UP000076502 UP000030765 UP000078492 UP000192223 UP000078541 UP000007755 UP000005205 UP000075884 UP000076407 UP000075903 UP000007062 UP000075840 UP000078540 UP000005203 UP000053825 UP000027135 UP000279307 UP000053097 UP000268350 UP000002358 UP000192221 UP000007798 UP000183832 UP000008744 UP000001819 UP000007801 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000092553 UP000079169 UP000001070 UP000008792 UP000009192 UP000015102 UP000095301 UP000092444 UP000037069
PRIDE
Pfam
PF05920 Homeobox_KN
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
A0A2W1BNZ6
A0A2A4JJ46
A0A2A4JKQ6
A0A139WL71
U4UQ21
N6U4R4
+ More
A0A1Y1LTB9 A0A1B0EWI7 A0A1S4FGP2 Q171W7 B0VZ28 A0A182GVJ9 A0A2M4CRT0 A0A2M4CRZ8 A0A154P8Q5 A0A084WDT6 A0A151J012 A0A1W4WZT5 A0A1W4WYS6 A0A1W4WP17 A0A195FNQ3 F4WMP6 A0A158N8X2 A0A1Y9H1X5 A0A2C9H895 A0A3F2Z0C0 Q7Q7U9 A0A2C9GPV8 A0A195B4P8 A0A1B6E4I8 A0A088AU13 A0A0C9RPT6 A0A0C9QIC1 A0A0L7QZW5 A0A067R8U0 A0A3L8DDD6 A0A2S2QBZ6 A0A026W5Y9 A0A3B0KMM3 K7IW51 A0A1W4UH91 B4NDK3 A0A1J1IJB5 A0A1B6GLA3 B4GRH6 A0A1B6M728 Q2M0K9 B3M9P9 B4HG50 B4QRQ3 O01667 Q9VU01 A0A0J9RVI5 B4PGT5 M9PI45 B3NCT4 A0A0M4EZX6 A0A1S4EBL4 B4IWZ1 B4LGH0 A0A0A9W3F2 W8B7E1 B4L0P7 A0A0A1WF40 A0A0K8UJ47 T1GN44 A0A1I8M686 A0A1I8M684 A0A336MWQ2 A0A336M4F8 A0A1B0FA50 A0A0L0C998 A0A0A9VZE3 A0A034VZD6 A0A2R7VVJ0
A0A1Y1LTB9 A0A1B0EWI7 A0A1S4FGP2 Q171W7 B0VZ28 A0A182GVJ9 A0A2M4CRT0 A0A2M4CRZ8 A0A154P8Q5 A0A084WDT6 A0A151J012 A0A1W4WZT5 A0A1W4WYS6 A0A1W4WP17 A0A195FNQ3 F4WMP6 A0A158N8X2 A0A1Y9H1X5 A0A2C9H895 A0A3F2Z0C0 Q7Q7U9 A0A2C9GPV8 A0A195B4P8 A0A1B6E4I8 A0A088AU13 A0A0C9RPT6 A0A0C9QIC1 A0A0L7QZW5 A0A067R8U0 A0A3L8DDD6 A0A2S2QBZ6 A0A026W5Y9 A0A3B0KMM3 K7IW51 A0A1W4UH91 B4NDK3 A0A1J1IJB5 A0A1B6GLA3 B4GRH6 A0A1B6M728 Q2M0K9 B3M9P9 B4HG50 B4QRQ3 O01667 Q9VU01 A0A0J9RVI5 B4PGT5 M9PI45 B3NCT4 A0A0M4EZX6 A0A1S4EBL4 B4IWZ1 B4LGH0 A0A0A9W3F2 W8B7E1 B4L0P7 A0A0A1WF40 A0A0K8UJ47 T1GN44 A0A1I8M686 A0A1I8M684 A0A336MWQ2 A0A336M4F8 A0A1B0FA50 A0A0L0C998 A0A0A9VZE3 A0A034VZD6 A0A2R7VVJ0
PDB
6FQP
E-value=1.06063e-07,
Score=134
Ontologies
GO
Topology
Subcellular location
Nucleus
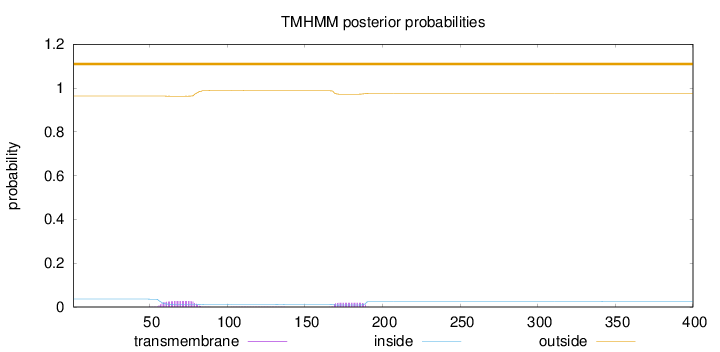
Length:
400
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.01635
Exp number, first 60 AAs:
0.0834
Total prob of N-in:
0.03612
outside
1 - 400
Population Genetic Test Statistics
Pi
18.186155
Theta
21.610092
Tajima's D
-2.186545
CLR
0.076017
CSRT
0.00574971251437428
Interpretation
Uncertain
