Gene
KWMTBOMO00217
Pre Gene Modal
BGIBMGA000574
Annotation
PREDICTED:_ankyrin_repeat_domain-containing_protein_SOWAHC-like_isoform_X4_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.045
Sequence
CDS
ATGCAGTATACTGCGATGCATTGGGCCTGTAAGCGAGGCGATGAAAATTTGGTGAAATTGCTAGGAGGAGTCGCTAGGCATGTTGTCAACGAACGTTCGGTTTTAAGTATCCTGTTAAAGTGTGATTACGTTATTTTTCATGTGGTGCCCGATACATGCCGTCGTCGATGGAAACAGAACGGGGGATACACGCCGCTACACATTGCTATGCAATTTCGACACGAGCATGTTTACCGGCTACTAGTCGAAGTATATGATGCAGATCCCAACATACGGGATTGGTCTGGAAGGAAACCAAGGCAATATTTAGTCCATATGGATACGTCATTATCGCCCGGCTCATACAGAAAAAAGGAGGGCTTCCTTCGTATCGGTTCGCTCAACGTTCGCGTTAAAAAAACAACGGAGGCCTTCAGTAATTTCCTCGGTGTTGGTGCGACGAGGTCGTCCGCGTACATGCCGAAATCGGCTCGGGAACGCGACGAAAGACGCTCCGACGACGGCGAACTTCACAAGACCTGGGGCTCTGCTGATAATATACAGAAGGATGACAAGTTAATGCCCCCGCCAATGAGTAGTAAAGTACGACGCAGGGGAGCCACGGGACGTCGCGGCGTAGGAGCTGCCCACAGTCGCAGCACTCCTTCCACGCCCGATCAGCCTCGAGCTCAGATGGGCGTCCCGGAAGAGAGAGATTCCGATTCTGATTCCGCAGCGGGATTCCATTCAGCATGGCGACAGCAACGTGCTTCAAACTGTACATAG
Protein
MQYTAMHWACKRGDENLVKLLGGVARHVVNERSVLSILLKCDYVIFHVVPDTCRRRWKQNGGYTPLHIAMQFRHEHVYRLLVEVYDADPNIRDWSGRKPRQYLVHMDTSLSPGSYRKKEGFLRIGSLNVRVKKTTEAFSNFLGVGATRSSAYMPKSARERDERRSDDGELHKTWGSADNIQKDDKLMPPPMSSKVRRRGATGRRGVGAAHSRSTPSTPDQPRAQMGVPEERDSDSDSAAGFHSAWRQQRASNCT
Summary
Uniprot
H9ITJ4
A0A2H1VUH1
A0A2A4J7S8
A0A212EMN0
A0A194QW18
A0A194PTC7
+ More
J9L4F6 A0A154P8Y4 A0A0A9X106 A0A151J030 A0A023EU59 A0A0A9WYF5 A0A1Q3FWW0 A0A1Y1L3U8 A0A0C9R8D4 A0A1Q3FX68 A0A158N8X1 A0A1B6H1K8 A0A0C9QXK2 E0W0Q8 A0A1Y1L6C3 A0A2C9H8A8 A0A2C9GPS3 A0A1S4GNF8 A0A1Y1L452 A0A2P8XXV0 A0A1B6KNZ5 A0A1B6LM90 A0A224XG55 A0A026W5Z9 A0A195CU83 K7IW53 A0A1B6LV74 A0A1B6E2K1 A0A1B6LXW9 A0A195B4X1 A0A1B6DDA7 A0A1B6CB35 A0A1Y1L7K5 A0A3L8DBS8 A0A0L7QZR6 A0A232FIZ4 A0A088ATZ9 A0A139WLJ4 E2AIB1 A0A336MNF3 Q2M0L3 A0A0R3P3K1 E2C0I2 F4WMN7 A0A195FQI9 B4IX66 A0A3B0KIZ6 A0A0J9RVH9 A0A1Y1L6B0 B4GRH1 A0A1S3DJ19 A0A1A9XGS5 A0A1D2MAD6 A0A182JJR9 A0A0P4Y8Z9 A0A0P6JU22 A0A0P5SIH7 A0A0P4W1G4 A0A0P5GGP7 A0A0M9A5M0 A0A182U4G7
J9L4F6 A0A154P8Y4 A0A0A9X106 A0A151J030 A0A023EU59 A0A0A9WYF5 A0A1Q3FWW0 A0A1Y1L3U8 A0A0C9R8D4 A0A1Q3FX68 A0A158N8X1 A0A1B6H1K8 A0A0C9QXK2 E0W0Q8 A0A1Y1L6C3 A0A2C9H8A8 A0A2C9GPS3 A0A1S4GNF8 A0A1Y1L452 A0A2P8XXV0 A0A1B6KNZ5 A0A1B6LM90 A0A224XG55 A0A026W5Z9 A0A195CU83 K7IW53 A0A1B6LV74 A0A1B6E2K1 A0A1B6LXW9 A0A195B4X1 A0A1B6DDA7 A0A1B6CB35 A0A1Y1L7K5 A0A3L8DBS8 A0A0L7QZR6 A0A232FIZ4 A0A088ATZ9 A0A139WLJ4 E2AIB1 A0A336MNF3 Q2M0L3 A0A0R3P3K1 E2C0I2 F4WMN7 A0A195FQI9 B4IX66 A0A3B0KIZ6 A0A0J9RVH9 A0A1Y1L6B0 B4GRH1 A0A1S3DJ19 A0A1A9XGS5 A0A1D2MAD6 A0A182JJR9 A0A0P4Y8Z9 A0A0P6JU22 A0A0P5SIH7 A0A0P4W1G4 A0A0P5GGP7 A0A0M9A5M0 A0A182U4G7
Pubmed
EMBL
BABH01006819
ODYU01004512
SOQ44470.1
NWSH01002669
PCG67718.1
AGBW02013811
+ More
OWR42760.1 KQ461108 KPJ09160.1 KQ459594 KPI96228.1 ABLF02024657 ABLF02024659 KQ434846 KZC08303.1 GBHO01031147 GBHO01031146 JAG12457.1 JAG12458.1 KQ980658 KYN14733.1 GAPW01000663 JAC12935.1 GBHO01031153 GBHO01031150 GBHO01031149 GBHO01031148 GBHO01016626 GBHO01016625 GBHO01016623 GBHO01016622 GBHO01016620 JAG12451.1 JAG12454.1 JAG12455.1 JAG12456.1 JAG26978.1 JAG26979.1 JAG26981.1 JAG26982.1 JAG26984.1 GFDL01002951 JAV32094.1 GEZM01065706 JAV68363.1 GBYB01012649 JAG82416.1 GFDL01002969 JAV32076.1 ADTU01008867 ADTU01008868 ADTU01008869 ADTU01008870 ADTU01008871 ADTU01008872 ADTU01008873 ADTU01008874 ADTU01008875 ADTU01008876 GECZ01001214 JAS68555.1 GBYB01005402 JAG75169.1 DS235862 EEB19214.1 GEZM01065708 JAV68361.1 APCN01004680 APCN01004681 AAAB01008948 GEZM01065705 JAV68364.1 PYGN01001183 PSN36825.1 GEBQ01026817 JAT13160.1 GEBQ01015174 JAT24803.1 GFTR01007628 JAW08798.1 KK107455 EZA50464.1 KQ977279 KYN04215.1 GEBQ01012386 JAT27591.1 GEDC01005146 JAS32152.1 GEBQ01011523 JAT28454.1 KQ976618 KYM79229.1 GEDC01013615 JAS23683.1 GEDC01026611 JAS10687.1 GEZM01065709 JAV68360.1 QOIP01000010 RLU17573.1 KQ414676 KOC64088.1 NNAY01000148 OXU30553.1 KQ971321 KYB28754.1 GL439754 EFN66822.1 UFQT01001628 SSX31235.1 CH379069 EAL30917.3 KRT07758.1 GL451797 EFN78541.1 GL888218 EGI64514.1 KQ981382 KYN42189.1 CH916366 EDV97398.1 OUUW01000009 SPP85061.1 CM002912 KMY99264.1 GEZM01065711 JAV68358.1 CH479188 EDW40356.1 LJIJ01002324 ODM89844.1 GDIP01234628 JAI88773.1 GDIQ01008368 JAN86369.1 GDIP01139340 JAL64374.1 GDRN01091941 JAI60197.1 GDIQ01241198 JAK10527.1 KQ435729 KOX77707.1
OWR42760.1 KQ461108 KPJ09160.1 KQ459594 KPI96228.1 ABLF02024657 ABLF02024659 KQ434846 KZC08303.1 GBHO01031147 GBHO01031146 JAG12457.1 JAG12458.1 KQ980658 KYN14733.1 GAPW01000663 JAC12935.1 GBHO01031153 GBHO01031150 GBHO01031149 GBHO01031148 GBHO01016626 GBHO01016625 GBHO01016623 GBHO01016622 GBHO01016620 JAG12451.1 JAG12454.1 JAG12455.1 JAG12456.1 JAG26978.1 JAG26979.1 JAG26981.1 JAG26982.1 JAG26984.1 GFDL01002951 JAV32094.1 GEZM01065706 JAV68363.1 GBYB01012649 JAG82416.1 GFDL01002969 JAV32076.1 ADTU01008867 ADTU01008868 ADTU01008869 ADTU01008870 ADTU01008871 ADTU01008872 ADTU01008873 ADTU01008874 ADTU01008875 ADTU01008876 GECZ01001214 JAS68555.1 GBYB01005402 JAG75169.1 DS235862 EEB19214.1 GEZM01065708 JAV68361.1 APCN01004680 APCN01004681 AAAB01008948 GEZM01065705 JAV68364.1 PYGN01001183 PSN36825.1 GEBQ01026817 JAT13160.1 GEBQ01015174 JAT24803.1 GFTR01007628 JAW08798.1 KK107455 EZA50464.1 KQ977279 KYN04215.1 GEBQ01012386 JAT27591.1 GEDC01005146 JAS32152.1 GEBQ01011523 JAT28454.1 KQ976618 KYM79229.1 GEDC01013615 JAS23683.1 GEDC01026611 JAS10687.1 GEZM01065709 JAV68360.1 QOIP01000010 RLU17573.1 KQ414676 KOC64088.1 NNAY01000148 OXU30553.1 KQ971321 KYB28754.1 GL439754 EFN66822.1 UFQT01001628 SSX31235.1 CH379069 EAL30917.3 KRT07758.1 GL451797 EFN78541.1 GL888218 EGI64514.1 KQ981382 KYN42189.1 CH916366 EDV97398.1 OUUW01000009 SPP85061.1 CM002912 KMY99264.1 GEZM01065711 JAV68358.1 CH479188 EDW40356.1 LJIJ01002324 ODM89844.1 GDIP01234628 JAI88773.1 GDIQ01008368 JAN86369.1 GDIP01139340 JAL64374.1 GDRN01091941 JAI60197.1 GDIQ01241198 JAK10527.1 KQ435729 KOX77707.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007819
+ More
UP000076502 UP000078492 UP000005205 UP000009046 UP000076407 UP000075840 UP000245037 UP000053097 UP000078542 UP000002358 UP000078540 UP000279307 UP000053825 UP000215335 UP000005203 UP000007266 UP000000311 UP000001819 UP000008237 UP000007755 UP000078541 UP000001070 UP000268350 UP000008744 UP000079169 UP000092443 UP000094527 UP000075880 UP000053105 UP000075902
UP000076502 UP000078492 UP000005205 UP000009046 UP000076407 UP000075840 UP000245037 UP000053097 UP000078542 UP000002358 UP000078540 UP000279307 UP000053825 UP000215335 UP000005203 UP000007266 UP000000311 UP000001819 UP000008237 UP000007755 UP000078541 UP000001070 UP000268350 UP000008744 UP000079169 UP000092443 UP000094527 UP000075880 UP000053105 UP000075902
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9ITJ4
A0A2H1VUH1
A0A2A4J7S8
A0A212EMN0
A0A194QW18
A0A194PTC7
+ More
J9L4F6 A0A154P8Y4 A0A0A9X106 A0A151J030 A0A023EU59 A0A0A9WYF5 A0A1Q3FWW0 A0A1Y1L3U8 A0A0C9R8D4 A0A1Q3FX68 A0A158N8X1 A0A1B6H1K8 A0A0C9QXK2 E0W0Q8 A0A1Y1L6C3 A0A2C9H8A8 A0A2C9GPS3 A0A1S4GNF8 A0A1Y1L452 A0A2P8XXV0 A0A1B6KNZ5 A0A1B6LM90 A0A224XG55 A0A026W5Z9 A0A195CU83 K7IW53 A0A1B6LV74 A0A1B6E2K1 A0A1B6LXW9 A0A195B4X1 A0A1B6DDA7 A0A1B6CB35 A0A1Y1L7K5 A0A3L8DBS8 A0A0L7QZR6 A0A232FIZ4 A0A088ATZ9 A0A139WLJ4 E2AIB1 A0A336MNF3 Q2M0L3 A0A0R3P3K1 E2C0I2 F4WMN7 A0A195FQI9 B4IX66 A0A3B0KIZ6 A0A0J9RVH9 A0A1Y1L6B0 B4GRH1 A0A1S3DJ19 A0A1A9XGS5 A0A1D2MAD6 A0A182JJR9 A0A0P4Y8Z9 A0A0P6JU22 A0A0P5SIH7 A0A0P4W1G4 A0A0P5GGP7 A0A0M9A5M0 A0A182U4G7
J9L4F6 A0A154P8Y4 A0A0A9X106 A0A151J030 A0A023EU59 A0A0A9WYF5 A0A1Q3FWW0 A0A1Y1L3U8 A0A0C9R8D4 A0A1Q3FX68 A0A158N8X1 A0A1B6H1K8 A0A0C9QXK2 E0W0Q8 A0A1Y1L6C3 A0A2C9H8A8 A0A2C9GPS3 A0A1S4GNF8 A0A1Y1L452 A0A2P8XXV0 A0A1B6KNZ5 A0A1B6LM90 A0A224XG55 A0A026W5Z9 A0A195CU83 K7IW53 A0A1B6LV74 A0A1B6E2K1 A0A1B6LXW9 A0A195B4X1 A0A1B6DDA7 A0A1B6CB35 A0A1Y1L7K5 A0A3L8DBS8 A0A0L7QZR6 A0A232FIZ4 A0A088ATZ9 A0A139WLJ4 E2AIB1 A0A336MNF3 Q2M0L3 A0A0R3P3K1 E2C0I2 F4WMN7 A0A195FQI9 B4IX66 A0A3B0KIZ6 A0A0J9RVH9 A0A1Y1L6B0 B4GRH1 A0A1S3DJ19 A0A1A9XGS5 A0A1D2MAD6 A0A182JJR9 A0A0P4Y8Z9 A0A0P6JU22 A0A0P5SIH7 A0A0P4W1G4 A0A0P5GGP7 A0A0M9A5M0 A0A182U4G7
PDB
4ZFH
E-value=0.00867445,
Score=90
Ontologies
GO
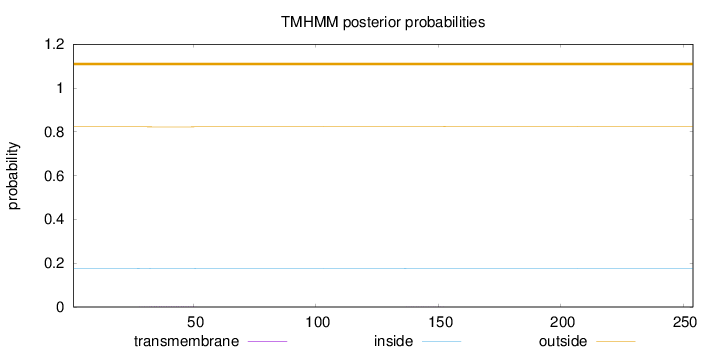
Topology
Length:
254
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0415100000000001
Exp number, first 60 AAs:
0.03835
Total prob of N-in:
0.17661
outside
1 - 254
Population Genetic Test Statistics
Pi
16.293771
Theta
23.399927
Tajima's D
-1.302739
CLR
0.02041
CSRT
0.0842957852107395
Interpretation
Uncertain
