Pre Gene Modal
BGIBMGA000581
Annotation
uncharacterized_protein_LOC101744278_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.841
Sequence
CDS
ATGGATGAAGAAGGCGAAAGCATTTTACTTGAAGATAATTATTATCAGCTTCTAAATGTATCAAAGACGGCTTCTTTAGAAGAAATCAATAGTGCATATCGTCGTTTCTCTCGAATGTTTCATCCAGACAAACATAGCACAGATCCCAACAAACAAAAATGGGCTGAGCAGATATTCAACAAAGTGAAAGAGGCTTATGAAGTTTTATCTGATTCACACAAAAGGGCCATATATGATACACTAGGAAAACGAGGTTTAGAAGTAGATGGTTGGGAGGTTATTTTCCGCACACGCACTCCTAGGGAAATAAGAGAAGAATATGAGAGGTTAAAACGGGAACGTGAGGAACGTAGGCTCCAGCAGAGTGCCAACCCTCGTGGCACAATCACACTCTCCATCAATGCCACAGATATGTTTACAAAGTATTATGATGAATATGAAATAATGGAAGAAACTACAGTGATCCCTAACATTGAAGTGTCAGGAATGACAATTCAACAATCCATTGATGCTCCAGTAACACTTCGAAATACAATGACTCTGTCAGGAAACATATCAACTCAGAATGGAATAGGTACTGGTTCAATAAATATGTGTAACAGGCACCTTAGCTCAGAAAAAGGCTGGACAGAAGTTGAATGTGGAATAGGAAATGGTCCTTTACTAGGTGTAAAATTATTTCGTACATTGTCTAGACTTATGTTTCTAAATTGTGGCACAGTGTTACAGTTTACATCAAGAGGAATTTCACCTAGCTTTGTTTCTACATTAGCTCTTCAGTTAGATGCTCACTCAGTGGGTTATTTGACATACCGAGCTGGAAACCAAGGCTCTTCAATGACTTCAATCTATGTGAGGGACTCGGAGAAATATCACAGCAACACTGCAATCCAGATTGGCAATCCGCATTCATTTATATCTTTCAATGTGATGAGGAAATTGCCCCAACATGATTTGAAGCTGAGACTGGCTGTAAAATTTGGGACATTTGGTGCAATAGCTGAGTATGGTGCGGAAAAGAAAGTGTCACAAAACAGTAGTGTGTCAGCTGCAGTTATGTTGGGTGTTCCTAGCGGGGTTATGCTCAAACTCAAGTGGACATGTTCATCACAAACTATAGTAGTGCCAATCCATCTGTGTGAGGAAGTGATGCCGTCACCAGTTTTCTACGCAACTGTAGTTCCATTAGTATCATGGATGATATTAAAAAAAATCGTTCTGGATCCTATTGCAAGAGAGCGTCAGGAACGAGAGAGACAACGTTCTATGGAAGCTAATTTTGAAAGATTACAGGAGATGCAAAGACAAGCACGCGCTACTGTAGAACTGATGAGAGAAACTTATTCCAGAATTCGGTCCCACGAAGATAAGAAGAAAGGTCTCGTCATATTAAAAGCTCTATACGGAAAGCTACCGGCAGATACATCGAGCCACGTGATACCCGAACAGACAGGAGACGGTGTATCTCCCGAATCGCCGAGTCCTTACAGCGATGTCATCGACGTCACAATACCCGTACAATGTTTGGTCAAAGATTCAAGGCTTGAACTTCTCGAGGCAAGCAAGTCGGAACTACCAGGCTTCTATGACCCGTGCGTTGGCGAGGACAAGCACTTGACAGTACAGTACATGTTTCACAATAATTTGCACTGCTGCACCGTACCTGACAATCAGGCCATCGTTCTGCCTCGCAACAATCATCGGATAAAGAATAGATCCTGA
Protein
MDEEGESILLEDNYYQLLNVSKTASLEEINSAYRRFSRMFHPDKHSTDPNKQKWAEQIFNKVKEAYEVLSDSHKRAIYDTLGKRGLEVDGWEVIFRTRTPREIREEYERLKREREERRLQQSANPRGTITLSINATDMFTKYYDEYEIMEETTVIPNIEVSGMTIQQSIDAPVTLRNTMTLSGNISTQNGIGTGSINMCNRHLSSEKGWTEVECGIGNGPLLGVKLFRTLSRLMFLNCGTVLQFTSRGISPSFVSTLALQLDAHSVGYLTYRAGNQGSSMTSIYVRDSEKYHSNTAIQIGNPHSFISFNVMRKLPQHDLKLRLAVKFGTFGAIAEYGAEKKVSQNSSVSAAVMLGVPSGVMLKLKWTCSSQTIVVPIHLCEEVMPSPVFYATVVPLVSWMILKKIVLDPIARERQERERQRSMEANFERLQEMQRQARATVELMRETYSRIRSHEDKKKGLVILKALYGKLPADTSSHVIPEQTGDGVSPESPSPYSDVIDVTIPVQCLVKDSRLELLEASKSELPGFYDPCVGEDKHLTVQYMFHNNLHCCTVPDNQAIVLPRNNHRIKNRS
Summary
Uniprot
H6VTQ7
A0A2A4K404
A0A194PSD0
A0A194QZZ9
A0A2H1V4N9
A0A212FMB0
+ More
A0A158NKC4 A0A195EHJ5 A0A067RFL1 A0A195FCD6 F4W8M4 A0A151I8Z1 E2AEG0 A0A026WN64 A0A2J7PJF1 A0A151I189 E9IBU1 A0A2J7PJG2 A0A151WTA2 A0A1B6D2D1 A0A0V0G382 A0A224XKU7 A0A0N7Z9K0 T1IBC2 A0A069DVH8 A0A0K8TKF3 A0A182GMZ2 Q16XI5 D6WKU6 A0A154PBL5 A0A2A3EF73 A0A087ZY83 A0A0T6B3Q9 A0A0K8TCV3 A0A1Q3FAI4 K7J700 A0A0A9YS62 B0WUV4 A0A232F5U5 A0A310S4U2 A0A182VDL6 A0A182HX94 A0A182WR93 A0A1S4H3Y8 A0A0J7KL42 A0A182RBD0 A0A182TV34 U5EWB7 A0A182LN96 E2C5N6 A0A2R7W5N6 Q7QDV2 N6SYM3 A0A0L7QT68 A0A336MG36 A0A084VZI2 A0A2M4AK11 W5J777 A0A182W3S8 A0A2M4BIW0 A0A182FM35 A0A182YSE0 A0A182QHK4 A0A182JFA2 A0A0M9A226 A0A182NDL1 A0A1Y1KEJ2 A0A1L8DU95 A0A182PVS3 A0A0C9QG58 A0A182JRE3 A0A0K8W4N6 A0A182SGW8 A0A034WPM1 A0A0P4XQA0 A0A0C9QP31 A0A0C9R6M4 A0A1I8M1N1 A0A0P6I1W9 A0A023GL89 A0A0P5WSP1 A0A0P5B563 A0A131XE83 A0A0P5BTG9 B3MDY6 A0A2P6KZX6 A0A131XYQ6 A0A1I8P8Z6 A0A131YNI3 L7M0I2 A0A224YLI0 A0A1E1X8U0 B3NR67 E9GBR0 A0A0L0CBQ2 W8CA82 A0A0P5DSJ7
A0A158NKC4 A0A195EHJ5 A0A067RFL1 A0A195FCD6 F4W8M4 A0A151I8Z1 E2AEG0 A0A026WN64 A0A2J7PJF1 A0A151I189 E9IBU1 A0A2J7PJG2 A0A151WTA2 A0A1B6D2D1 A0A0V0G382 A0A224XKU7 A0A0N7Z9K0 T1IBC2 A0A069DVH8 A0A0K8TKF3 A0A182GMZ2 Q16XI5 D6WKU6 A0A154PBL5 A0A2A3EF73 A0A087ZY83 A0A0T6B3Q9 A0A0K8TCV3 A0A1Q3FAI4 K7J700 A0A0A9YS62 B0WUV4 A0A232F5U5 A0A310S4U2 A0A182VDL6 A0A182HX94 A0A182WR93 A0A1S4H3Y8 A0A0J7KL42 A0A182RBD0 A0A182TV34 U5EWB7 A0A182LN96 E2C5N6 A0A2R7W5N6 Q7QDV2 N6SYM3 A0A0L7QT68 A0A336MG36 A0A084VZI2 A0A2M4AK11 W5J777 A0A182W3S8 A0A2M4BIW0 A0A182FM35 A0A182YSE0 A0A182QHK4 A0A182JFA2 A0A0M9A226 A0A182NDL1 A0A1Y1KEJ2 A0A1L8DU95 A0A182PVS3 A0A0C9QG58 A0A182JRE3 A0A0K8W4N6 A0A182SGW8 A0A034WPM1 A0A0P4XQA0 A0A0C9QP31 A0A0C9R6M4 A0A1I8M1N1 A0A0P6I1W9 A0A023GL89 A0A0P5WSP1 A0A0P5B563 A0A131XE83 A0A0P5BTG9 B3MDY6 A0A2P6KZX6 A0A131XYQ6 A0A1I8P8Z6 A0A131YNI3 L7M0I2 A0A224YLI0 A0A1E1X8U0 B3NR67 E9GBR0 A0A0L0CBQ2 W8CA82 A0A0P5DSJ7
Pubmed
26434795
26354079
22118469
21347285
24845553
21719571
+ More
20798317 24508170 30249741 21282665 27129103 26334808 26369729 26483478 17510324 18362917 19820115 26823975 20075255 25401762 28648823 12364791 20966253 23537049 24438588 20920257 23761445 25244985 28004739 25348373 25315136 17994087 18057021 26830274 25576852 28797301 28503490 21292972 26108605 24495485
20798317 24508170 30249741 21282665 27129103 26334808 26369729 26483478 17510324 18362917 19820115 26823975 20075255 25401762 28648823 12364791 20966253 23537049 24438588 20920257 23761445 25244985 28004739 25348373 25315136 17994087 18057021 26830274 25576852 28797301 28503490 21292972 26108605 24495485
EMBL
JN872892
AFC01231.1
NWSH01000187
PCG78634.1
KQ459594
KPI96217.1
+ More
KQ461108 KPJ09171.1 ODYU01000482 SOQ35342.1 AGBW02007654 OWR54867.1 ADTU01018770 KQ978881 KYN27745.1 KK852665 KDR18967.1 KQ981693 KYN37704.1 GL887908 EGI69515.1 KQ978323 KYM95123.1 GL438838 EFN68171.1 KK107148 QOIP01000009 EZA57398.1 RLU18320.1 NEVH01024949 PNF16475.1 KQ976588 KYM79697.1 GL762137 EFZ21995.1 PNF16476.1 KQ982762 KYQ51021.1 GEDC01017492 JAS19806.1 GECL01003574 JAP02550.1 GFTR01007204 JAW09222.1 GDKW01000331 JAI56264.1 ACPB03000856 GBGD01001004 JAC87885.1 GDAI01002776 JAI14827.1 JXUM01075431 JXUM01075432 JXUM01075433 KQ562886 KXJ74917.1 CH477538 EAT39324.1 KQ971343 EFA04019.2 KQ434868 KZC09223.1 KZ288266 PBC30134.1 LJIG01016002 KRT81908.1 GBRD01002429 GDHC01000257 JAG63392.1 JAQ18372.1 GFDL01010459 JAV24586.1 GBHO01008580 JAG35024.1 DS232111 EDS35097.1 NNAY01000900 OXU26015.1 KQ773953 OAD52344.1 APCN01005160 AAAB01008849 LBMM01006080 KMQ90951.1 GANO01001492 JAB58379.1 GL452776 EFN76747.1 KK854357 PTY15046.1 EAA07236.4 APGK01051809 APGK01051810 KB741207 ENN72879.1 KQ414756 KOC61671.1 UFQT01001098 SSX28900.1 ATLV01018848 KE525251 KFB43376.1 GGFK01007637 MBW40958.1 ADMH02002126 ETN58639.1 GGFJ01003865 MBW53006.1 AXCN02001753 KQ435760 KOX75607.1 GEZM01088910 JAV57836.1 GFDF01004087 JAV09997.1 GBYB01002419 JAG72186.1 GDHF01006489 JAI45825.1 GAKP01002363 JAC56589.1 GDIP01238280 JAI85121.1 GBYB01002417 JAG72184.1 GBYB01002416 JAG72183.1 GDIQ01010712 LRGB01002860 JAN84025.1 KZS05849.1 GBBM01000772 JAC34646.1 GDIP01094601 JAM09114.1 GDIP01189479 JAJ33923.1 GEFH01004096 JAP64485.1 GDIP01180130 JAJ43272.1 CH902619 EDV37531.1 KPU76892.1 MWRG01003124 PRD31873.1 GEFM01003433 JAP72363.1 GEDV01008040 JAP80517.1 GACK01007652 JAA57382.1 GFPF01007352 MAA18498.1 GFAC01003544 JAT95644.1 CH954179 EDV56056.1 GL732538 EFX83114.1 JRES01000644 KNC29667.1 GAMC01001514 JAC05042.1 GDIP01152292 JAJ71110.1
KQ461108 KPJ09171.1 ODYU01000482 SOQ35342.1 AGBW02007654 OWR54867.1 ADTU01018770 KQ978881 KYN27745.1 KK852665 KDR18967.1 KQ981693 KYN37704.1 GL887908 EGI69515.1 KQ978323 KYM95123.1 GL438838 EFN68171.1 KK107148 QOIP01000009 EZA57398.1 RLU18320.1 NEVH01024949 PNF16475.1 KQ976588 KYM79697.1 GL762137 EFZ21995.1 PNF16476.1 KQ982762 KYQ51021.1 GEDC01017492 JAS19806.1 GECL01003574 JAP02550.1 GFTR01007204 JAW09222.1 GDKW01000331 JAI56264.1 ACPB03000856 GBGD01001004 JAC87885.1 GDAI01002776 JAI14827.1 JXUM01075431 JXUM01075432 JXUM01075433 KQ562886 KXJ74917.1 CH477538 EAT39324.1 KQ971343 EFA04019.2 KQ434868 KZC09223.1 KZ288266 PBC30134.1 LJIG01016002 KRT81908.1 GBRD01002429 GDHC01000257 JAG63392.1 JAQ18372.1 GFDL01010459 JAV24586.1 GBHO01008580 JAG35024.1 DS232111 EDS35097.1 NNAY01000900 OXU26015.1 KQ773953 OAD52344.1 APCN01005160 AAAB01008849 LBMM01006080 KMQ90951.1 GANO01001492 JAB58379.1 GL452776 EFN76747.1 KK854357 PTY15046.1 EAA07236.4 APGK01051809 APGK01051810 KB741207 ENN72879.1 KQ414756 KOC61671.1 UFQT01001098 SSX28900.1 ATLV01018848 KE525251 KFB43376.1 GGFK01007637 MBW40958.1 ADMH02002126 ETN58639.1 GGFJ01003865 MBW53006.1 AXCN02001753 KQ435760 KOX75607.1 GEZM01088910 JAV57836.1 GFDF01004087 JAV09997.1 GBYB01002419 JAG72186.1 GDHF01006489 JAI45825.1 GAKP01002363 JAC56589.1 GDIP01238280 JAI85121.1 GBYB01002417 JAG72184.1 GBYB01002416 JAG72183.1 GDIQ01010712 LRGB01002860 JAN84025.1 KZS05849.1 GBBM01000772 JAC34646.1 GDIP01094601 JAM09114.1 GDIP01189479 JAJ33923.1 GEFH01004096 JAP64485.1 GDIP01180130 JAJ43272.1 CH902619 EDV37531.1 KPU76892.1 MWRG01003124 PRD31873.1 GEFM01003433 JAP72363.1 GEDV01008040 JAP80517.1 GACK01007652 JAA57382.1 GFPF01007352 MAA18498.1 GFAC01003544 JAT95644.1 CH954179 EDV56056.1 GL732538 EFX83114.1 JRES01000644 KNC29667.1 GAMC01001514 JAC05042.1 GDIP01152292 JAJ71110.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000007151
UP000005205
UP000078492
+ More
UP000027135 UP000078541 UP000007755 UP000078542 UP000000311 UP000053097 UP000279307 UP000235965 UP000078540 UP000075809 UP000015103 UP000069940 UP000249989 UP000008820 UP000007266 UP000076502 UP000242457 UP000005203 UP000002358 UP000002320 UP000215335 UP000075903 UP000075840 UP000076407 UP000036403 UP000075900 UP000075902 UP000075882 UP000008237 UP000007062 UP000019118 UP000053825 UP000030765 UP000000673 UP000075920 UP000069272 UP000076408 UP000075886 UP000075880 UP000053105 UP000075884 UP000075885 UP000075881 UP000075901 UP000095301 UP000076858 UP000007801 UP000095300 UP000008711 UP000000305 UP000037069
UP000027135 UP000078541 UP000007755 UP000078542 UP000000311 UP000053097 UP000279307 UP000235965 UP000078540 UP000075809 UP000015103 UP000069940 UP000249989 UP000008820 UP000007266 UP000076502 UP000242457 UP000005203 UP000002358 UP000002320 UP000215335 UP000075903 UP000075840 UP000076407 UP000036403 UP000075900 UP000075902 UP000075882 UP000008237 UP000007062 UP000019118 UP000053825 UP000030765 UP000000673 UP000075920 UP000069272 UP000076408 UP000075886 UP000075880 UP000053105 UP000075884 UP000075885 UP000075881 UP000075901 UP000095301 UP000076858 UP000007801 UP000095300 UP000008711 UP000000305 UP000037069
PRIDE
Interpro
SUPFAM
SSF46565
SSF46565
Gene 3D
CDD
ProteinModelPortal
H6VTQ7
A0A2A4K404
A0A194PSD0
A0A194QZZ9
A0A2H1V4N9
A0A212FMB0
+ More
A0A158NKC4 A0A195EHJ5 A0A067RFL1 A0A195FCD6 F4W8M4 A0A151I8Z1 E2AEG0 A0A026WN64 A0A2J7PJF1 A0A151I189 E9IBU1 A0A2J7PJG2 A0A151WTA2 A0A1B6D2D1 A0A0V0G382 A0A224XKU7 A0A0N7Z9K0 T1IBC2 A0A069DVH8 A0A0K8TKF3 A0A182GMZ2 Q16XI5 D6WKU6 A0A154PBL5 A0A2A3EF73 A0A087ZY83 A0A0T6B3Q9 A0A0K8TCV3 A0A1Q3FAI4 K7J700 A0A0A9YS62 B0WUV4 A0A232F5U5 A0A310S4U2 A0A182VDL6 A0A182HX94 A0A182WR93 A0A1S4H3Y8 A0A0J7KL42 A0A182RBD0 A0A182TV34 U5EWB7 A0A182LN96 E2C5N6 A0A2R7W5N6 Q7QDV2 N6SYM3 A0A0L7QT68 A0A336MG36 A0A084VZI2 A0A2M4AK11 W5J777 A0A182W3S8 A0A2M4BIW0 A0A182FM35 A0A182YSE0 A0A182QHK4 A0A182JFA2 A0A0M9A226 A0A182NDL1 A0A1Y1KEJ2 A0A1L8DU95 A0A182PVS3 A0A0C9QG58 A0A182JRE3 A0A0K8W4N6 A0A182SGW8 A0A034WPM1 A0A0P4XQA0 A0A0C9QP31 A0A0C9R6M4 A0A1I8M1N1 A0A0P6I1W9 A0A023GL89 A0A0P5WSP1 A0A0P5B563 A0A131XE83 A0A0P5BTG9 B3MDY6 A0A2P6KZX6 A0A131XYQ6 A0A1I8P8Z6 A0A131YNI3 L7M0I2 A0A224YLI0 A0A1E1X8U0 B3NR67 E9GBR0 A0A0L0CBQ2 W8CA82 A0A0P5DSJ7
A0A158NKC4 A0A195EHJ5 A0A067RFL1 A0A195FCD6 F4W8M4 A0A151I8Z1 E2AEG0 A0A026WN64 A0A2J7PJF1 A0A151I189 E9IBU1 A0A2J7PJG2 A0A151WTA2 A0A1B6D2D1 A0A0V0G382 A0A224XKU7 A0A0N7Z9K0 T1IBC2 A0A069DVH8 A0A0K8TKF3 A0A182GMZ2 Q16XI5 D6WKU6 A0A154PBL5 A0A2A3EF73 A0A087ZY83 A0A0T6B3Q9 A0A0K8TCV3 A0A1Q3FAI4 K7J700 A0A0A9YS62 B0WUV4 A0A232F5U5 A0A310S4U2 A0A182VDL6 A0A182HX94 A0A182WR93 A0A1S4H3Y8 A0A0J7KL42 A0A182RBD0 A0A182TV34 U5EWB7 A0A182LN96 E2C5N6 A0A2R7W5N6 Q7QDV2 N6SYM3 A0A0L7QT68 A0A336MG36 A0A084VZI2 A0A2M4AK11 W5J777 A0A182W3S8 A0A2M4BIW0 A0A182FM35 A0A182YSE0 A0A182QHK4 A0A182JFA2 A0A0M9A226 A0A182NDL1 A0A1Y1KEJ2 A0A1L8DU95 A0A182PVS3 A0A0C9QG58 A0A182JRE3 A0A0K8W4N6 A0A182SGW8 A0A034WPM1 A0A0P4XQA0 A0A0C9QP31 A0A0C9R6M4 A0A1I8M1N1 A0A0P6I1W9 A0A023GL89 A0A0P5WSP1 A0A0P5B563 A0A131XE83 A0A0P5BTG9 B3MDY6 A0A2P6KZX6 A0A131XYQ6 A0A1I8P8Z6 A0A131YNI3 L7M0I2 A0A224YLI0 A0A1E1X8U0 B3NR67 E9GBR0 A0A0L0CBQ2 W8CA82 A0A0P5DSJ7
PDB
2LGW
E-value=3.36317e-15,
Score=201
Ontologies
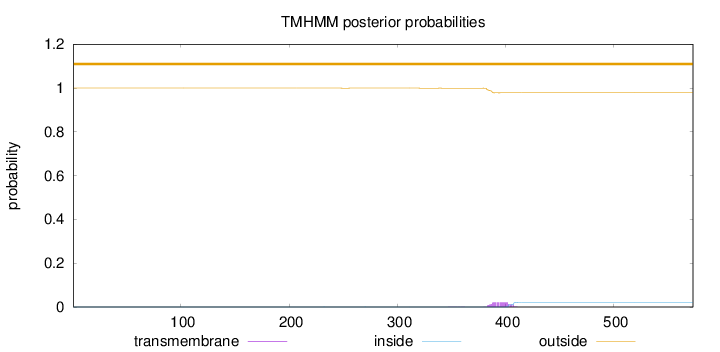
Topology
Length:
573
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.44775
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00036
outside
1 - 573
Population Genetic Test Statistics
Pi
1.802154
Theta
21.779138
Tajima's D
-2.401013
CLR
0.149551
CSRT
0.00134993250337483
Interpretation
Uncertain
