Gene
KWMTBOMO00209
Pre Gene Modal
BGIBMGA000582
Annotation
PREDICTED:_UNC93-like_protein_[Amyelois_transitella]
Full name
UNC93-like protein
Location in the cell
PlasmaMembrane Reliability : 4.617
Sequence
CDS
ATGACGGCTACTCACGGAGGCGGACGTGAAGATGAACGTCCGGACAAAGATTCTGTATACACCATCAAAATCGGGTTCAAGAATGATGGTTATCAGCACGACCGCGACTCGAACGATGACATAGTCAAACCCCCGCCTTTGCCCACCGAGGAGGACAACTACTCAACCGGAAAAGTAAAGCTTTCCAGGAACGAGAAATGGAGAATTCTAAAGAACGTGGCAGCGGTTAGCGCCGCATTCATGGTCCAGTTCACCGCTTTCCAGGGAACCGCAAACCTGCAATCGTCGATAAACGCCGCAGATGGCCTAGGAACAGTCTCACTGAGTTCCATCTACGCAGCCTTGGTAGTCTCGTGTATATTTGTACCAACGTTCCTGATAAAAAGACTAACTGTTAAATGGACCCTTTGCCTCTCAATGATATGTTACGCACCATACATCGGGGCCCAGTTCTACCCGGCATTCTACACGCTGGTGCCGGCGGGTGTTATTGTAGGTCTGGGCGCAGCTCCTATGTGGACTGCAAAAGCGACGTATCTCACGCAAGCGGGTAGCGTTTACGCGAAGCTAACTGACCAAGCCGTTGACGGGATTATCGTGCGATTCTTCGGCTTCTTCTTCCTAGCTTGGCAAACGGCTGAACTGTGGGGAAATCTGATCTCAAGCCTCGTGTTTTCATCGGGAGAGCACAGCACAAACTCTACAGTCGTGGATACCAACAAGACTGCCCTTACGTGTGGAGCTAACTTTTGCGTCATAGGAGGTGGTCACCATGACAACCAGAATTTGCACCGACCACCGGACAGCGAGATCTACGAAATCAGTGCAATTTACTTATCCTGTGTCGTTGTCGCCGTCATAATGGTAGCCCTTCTTGTTGACCCATTGTCTAGGTATGGTGAAAAACAAAGGAAAGCTGATCCATCTAAGGAGCTCAGCGGTATACAATTGTTATCTGCTACCGCTTATCAGCTGAAGAAGCCAAATCAGCAACTTCTTATTCCAATCACCATCTGGATTGGAATGGAGCAAGCTTTTATTGGAGCTGACTACACTCAGGCATACGTCTCGTGCGTTCTCGGAATTCGCTCTATAGGCTACGTCATGATATGCTTCGGCGTCGTCAACGCTCTCTGTTCGCTGGTATTTGGTTCGGCCATGAAGTTCATCGGTAGATTTCCCATATTGATAATGGGGGCCGCACTGCATTTCGGCCTGATTGTCTGGCTGCTTGTATGGAAACCCGACCCCAGATTTCCGACGGTGTTCTTCGTAATTTCTGGACTGTGGGGTGTTGGAGATGCCGTATGGCAAACTCAAGTCAACGGCTTGTACGGTGTCCTGTTCAGGCGCAACAAAGAGGCTGCGTTTTCAAACTACCGCCTCTGGGAAGGCGCTGGTTTCGTTGTGGCTTATGCCTACTCAACGCACCTGTGCGCCAGAATGAAGCTTTACGTGATGATGGTGGTGCTCTTAATCGGTGTCGTTGGATACATTATAGTTGAAATTCTACATAAAAGGAAGGCTCTTCGTCAGAAGGCTATCGCAGAGGACCCGGCAATCGCCGCTCAAGCCGCCAAAGAACCACCAGAAGAAGACGACGAGAAGGATGAAATTGACGACGAAATCGTCATCACTCACCTGTAA
Protein
MTATHGGGREDERPDKDSVYTIKIGFKNDGYQHDRDSNDDIVKPPPLPTEEDNYSTGKVKLSRNEKWRILKNVAAVSAAFMVQFTAFQGTANLQSSINAADGLGTVSLSSIYAALVVSCIFVPTFLIKRLTVKWTLCLSMICYAPYIGAQFYPAFYTLVPAGVIVGLGAAPMWTAKATYLTQAGSVYAKLTDQAVDGIIVRFFGFFFLAWQTAELWGNLISSLVFSSGEHSTNSTVVDTNKTALTCGANFCVIGGGHHDNQNLHRPPDSEIYEISAIYLSCVVVAVIMVALLVDPLSRYGEKQRKADPSKELSGIQLLSATAYQLKKPNQQLLIPITIWIGMEQAFIGADYTQAYVSCVLGIRSIGYVMICFGVVNALCSLVFGSAMKFIGRFPILIMGAALHFGLIVWLLVWKPDPRFPTVFFVISGLWGVGDAVWQTQVNGLYGVLFRRNKEAAFSNYRLWEGAGFVVAYAYSTHLCARMKLYVMMVVLLIGVVGYIIVEILHKRKALRQKAIAEDPAIAAQAAKEPPEEDDEKDEIDDEIVITHL
Summary
Similarity
Belongs to the unc-93 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain UNC93-like protein
Uniprot
H9ITK2
A0A2A4K4E7
A0A2W1BNK6
A0A194PYI7
A0A194QUG9
A0A212FMF8
+ More
A0A2A3EB67 A0A088A7F7 A0A195CTX8 A0A151JZ88 A0A158NFD5 A0A1B6CT84 A0A1B6DYS0 A0A1B6CEC5 A0A023F4B7 A0A3L8DGP3 A0A0C9Q4D2 A0A0L7LTD7 K7IW01 A0A069DZ97 J9JT06 A0A0M8ZRY9 A0A0J7L5W3 A0A1Y1NB36 T1I6X0 A0A151XFM2 A0A1Y1NB73 A0A2S2Q6F7 A0A1B6M7T8 A0A310SRE3 A0A2H8TF89 A0A2S2NP74 A0A1B6G7H6 A0A1W4WFI9 A0A1W4W3P0 A0A1W4WE96 A0A1W4W3P9 A0A139WL24 A0A2J7QTX9 V5I8T1 A0A182G9R9 A0A146M0F1 B0W7P2 A0A182GWR4 A0A1Q3FR00 A0A182JCV9 Q17DS4 A0A182XXX1 A0A084VQ76 E0W1U1 A0A182QC59 A0A2P8YI88 A0A182UV07 A0A232F9S6 A0A034W050 A0A182KH76 A0A182FIU1 W8AD07 A0A0A9YMX1 Q7PNJ1 F5HLY6 A0A0K8V7B1 A0A0A1WEG1 A0A182NV40 A0A182RMU1 N6TTC5 W5JS93 A0A1I8PPG6 A0A034W1Q1 A0A336LQI1 T1PC15 B4JN53 A0A0A1XKD8 Q6BCZ5 A0A0L0C1N1 A0A1J1IIF3 A0A1A9YQC3 A0A1L8D975 B4M1K1 A0A182SBW0 B4L229 X2JFT8 A0A0M4ELF5 B7Z155 B3NWA1 B4Q1X3 Q9Y115 B4R656 B4IEV4 B3N0B8 B4HA36 Q29FL5 Q6BCZ8 Q6BD00 Q6BCZ7 Q6BCZ1 A0A1A9WHR3 A0A1W4VWV5 A0A3B0J7N1
A0A2A3EB67 A0A088A7F7 A0A195CTX8 A0A151JZ88 A0A158NFD5 A0A1B6CT84 A0A1B6DYS0 A0A1B6CEC5 A0A023F4B7 A0A3L8DGP3 A0A0C9Q4D2 A0A0L7LTD7 K7IW01 A0A069DZ97 J9JT06 A0A0M8ZRY9 A0A0J7L5W3 A0A1Y1NB36 T1I6X0 A0A151XFM2 A0A1Y1NB73 A0A2S2Q6F7 A0A1B6M7T8 A0A310SRE3 A0A2H8TF89 A0A2S2NP74 A0A1B6G7H6 A0A1W4WFI9 A0A1W4W3P0 A0A1W4WE96 A0A1W4W3P9 A0A139WL24 A0A2J7QTX9 V5I8T1 A0A182G9R9 A0A146M0F1 B0W7P2 A0A182GWR4 A0A1Q3FR00 A0A182JCV9 Q17DS4 A0A182XXX1 A0A084VQ76 E0W1U1 A0A182QC59 A0A2P8YI88 A0A182UV07 A0A232F9S6 A0A034W050 A0A182KH76 A0A182FIU1 W8AD07 A0A0A9YMX1 Q7PNJ1 F5HLY6 A0A0K8V7B1 A0A0A1WEG1 A0A182NV40 A0A182RMU1 N6TTC5 W5JS93 A0A1I8PPG6 A0A034W1Q1 A0A336LQI1 T1PC15 B4JN53 A0A0A1XKD8 Q6BCZ5 A0A0L0C1N1 A0A1J1IIF3 A0A1A9YQC3 A0A1L8D975 B4M1K1 A0A182SBW0 B4L229 X2JFT8 A0A0M4ELF5 B7Z155 B3NWA1 B4Q1X3 Q9Y115 B4R656 B4IEV4 B3N0B8 B4HA36 Q29FL5 Q6BCZ8 Q6BD00 Q6BCZ7 Q6BCZ1 A0A1A9WHR3 A0A1W4VWV5 A0A3B0J7N1
Pubmed
19121390
28756777
26354079
22118469
21347285
25474469
+ More
30249741 26227816 20075255 26334808 28004739 18362917 19820115 26483478 26823975 17510324 25244985 24438588 20566863 29403074 28648823 25348373 24495485 25401762 12364791 14747013 17210077 25830018 23537049 20920257 23761445 25315136 17994087 15579698 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 10731138 15632085 23185243
30249741 26227816 20075255 26334808 28004739 18362917 19820115 26483478 26823975 17510324 25244985 24438588 20566863 29403074 28648823 25348373 24495485 25401762 12364791 14747013 17210077 25830018 23537049 20920257 23761445 25315136 17994087 15579698 26108605 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 10731138 15632085 23185243
EMBL
BABH01006778
NWSH01000187
PCG78633.1
KZ150006
PZC75205.1
KQ459594
+ More
KPI96215.1 KQ461108 KPJ09173.1 AGBW02007654 OWR54869.1 KZ288296 PBC28930.1 KQ977279 KYN03977.1 KQ981410 KYN41916.1 ADTU01014275 ADTU01014276 ADTU01014277 ADTU01014278 GEDC01020618 JAS16680.1 GEDC01006499 JAS30799.1 GEDC01025529 JAS11769.1 GBBI01002361 JAC16351.1 QOIP01000009 RLU19038.1 GBYB01008918 JAG78685.1 JTDY01000125 KOB78720.1 GBGD01001020 JAC87869.1 ABLF02027629 ABLF02027631 KQ435918 KOX68736.1 LBMM01000627 KMQ97943.1 GEZM01007503 GEZM01007501 JAV95142.1 ACPB03000765 KQ982182 KYQ59183.1 GEZM01007502 JAV95141.1 GGMS01004123 MBY73326.1 GEBQ01007982 JAT31995.1 KQ759878 OAD62254.1 GFXV01000936 MBW12741.1 GGMR01006335 MBY18954.1 GECZ01011381 JAS58388.1 KQ971324 KYB28531.1 NEVH01011192 PNF32038.1 GALX01004145 JAB64321.1 JXUM01049812 KQ561622 KXJ77978.1 GDHC01006549 JAQ12080.1 DS231855 EDS38186.1 JXUM01003156 KQ560154 KXJ84146.1 GFDL01004994 JAV30051.1 CH477291 EAT44590.1 ATLV01015142 KE525003 KFB40120.1 DS235873 EEB19673.1 AXCN02001448 AXCN02001449 PYGN01000576 PSN43975.1 NNAY01000574 OXU27596.1 GAKP01011270 JAC47682.1 GAMC01020698 GAMC01020695 JAB85860.1 GBHO01009202 GBRD01002011 GBRD01002009 JAG34402.1 JAG63810.1 AAAB01008963 EAA12091.6 EGK97297.1 GDHF01017512 JAI34802.1 GBXI01017509 JAC96782.1 APGK01019729 APGK01019730 KB740120 ENN81323.1 ADMH02000612 ETN65624.1 GAKP01011269 JAC47683.1 UFQT01000116 SSX20314.1 KA645473 AFP60102.1 CH916371 EDV92146.1 GBXI01002962 JAD11330.1 AY665388 AAT76555.1 JRES01001003 KNC26210.1 CVRI01000048 CRK98846.1 GFDF01011081 JAV03003.1 CH940651 EDW65555.1 KRF82281.1 CH933810 EDW06769.1 KRF93808.1 KRF93809.1 AE014298 AHN59824.1 CP012528 ALC48271.1 AAF48682.1 CH954180 EDV46440.1 KQS29972.1 KQS29973.1 CM000162 EDX02548.1 KRK06824.1 KRK06825.1 AF145657 AY665384 CM000366 EDX18160.1 CH480832 EDW46208.1 CH902640 EDV38322.1 KPU77362.1 CH479235 EDW36704.1 CH379066 EAL31385.1 KRT07333.1 AY665386 AAT76553.1 AY665385 AAT76552.1 AY665387 AAT76554.1 AY665390 AAT76557.1 OUUW01000003 SPP78117.1
KPI96215.1 KQ461108 KPJ09173.1 AGBW02007654 OWR54869.1 KZ288296 PBC28930.1 KQ977279 KYN03977.1 KQ981410 KYN41916.1 ADTU01014275 ADTU01014276 ADTU01014277 ADTU01014278 GEDC01020618 JAS16680.1 GEDC01006499 JAS30799.1 GEDC01025529 JAS11769.1 GBBI01002361 JAC16351.1 QOIP01000009 RLU19038.1 GBYB01008918 JAG78685.1 JTDY01000125 KOB78720.1 GBGD01001020 JAC87869.1 ABLF02027629 ABLF02027631 KQ435918 KOX68736.1 LBMM01000627 KMQ97943.1 GEZM01007503 GEZM01007501 JAV95142.1 ACPB03000765 KQ982182 KYQ59183.1 GEZM01007502 JAV95141.1 GGMS01004123 MBY73326.1 GEBQ01007982 JAT31995.1 KQ759878 OAD62254.1 GFXV01000936 MBW12741.1 GGMR01006335 MBY18954.1 GECZ01011381 JAS58388.1 KQ971324 KYB28531.1 NEVH01011192 PNF32038.1 GALX01004145 JAB64321.1 JXUM01049812 KQ561622 KXJ77978.1 GDHC01006549 JAQ12080.1 DS231855 EDS38186.1 JXUM01003156 KQ560154 KXJ84146.1 GFDL01004994 JAV30051.1 CH477291 EAT44590.1 ATLV01015142 KE525003 KFB40120.1 DS235873 EEB19673.1 AXCN02001448 AXCN02001449 PYGN01000576 PSN43975.1 NNAY01000574 OXU27596.1 GAKP01011270 JAC47682.1 GAMC01020698 GAMC01020695 JAB85860.1 GBHO01009202 GBRD01002011 GBRD01002009 JAG34402.1 JAG63810.1 AAAB01008963 EAA12091.6 EGK97297.1 GDHF01017512 JAI34802.1 GBXI01017509 JAC96782.1 APGK01019729 APGK01019730 KB740120 ENN81323.1 ADMH02000612 ETN65624.1 GAKP01011269 JAC47683.1 UFQT01000116 SSX20314.1 KA645473 AFP60102.1 CH916371 EDV92146.1 GBXI01002962 JAD11330.1 AY665388 AAT76555.1 JRES01001003 KNC26210.1 CVRI01000048 CRK98846.1 GFDF01011081 JAV03003.1 CH940651 EDW65555.1 KRF82281.1 CH933810 EDW06769.1 KRF93808.1 KRF93809.1 AE014298 AHN59824.1 CP012528 ALC48271.1 AAF48682.1 CH954180 EDV46440.1 KQS29972.1 KQS29973.1 CM000162 EDX02548.1 KRK06824.1 KRK06825.1 AF145657 AY665384 CM000366 EDX18160.1 CH480832 EDW46208.1 CH902640 EDV38322.1 KPU77362.1 CH479235 EDW36704.1 CH379066 EAL31385.1 KRT07333.1 AY665386 AAT76553.1 AY665385 AAT76552.1 AY665387 AAT76554.1 AY665390 AAT76557.1 OUUW01000003 SPP78117.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000242457
+ More
UP000005203 UP000078542 UP000078541 UP000005205 UP000279307 UP000037510 UP000002358 UP000007819 UP000053105 UP000036403 UP000015103 UP000075809 UP000192223 UP000007266 UP000235965 UP000069940 UP000249989 UP000002320 UP000075880 UP000008820 UP000076408 UP000030765 UP000009046 UP000075886 UP000245037 UP000075903 UP000215335 UP000075881 UP000069272 UP000007062 UP000075884 UP000075900 UP000019118 UP000000673 UP000095300 UP000095301 UP000001070 UP000037069 UP000183832 UP000092443 UP000008792 UP000075901 UP000009192 UP000000803 UP000092553 UP000008711 UP000002282 UP000000304 UP000001292 UP000007801 UP000008744 UP000001819 UP000091820 UP000192221 UP000268350
UP000005203 UP000078542 UP000078541 UP000005205 UP000279307 UP000037510 UP000002358 UP000007819 UP000053105 UP000036403 UP000015103 UP000075809 UP000192223 UP000007266 UP000235965 UP000069940 UP000249989 UP000002320 UP000075880 UP000008820 UP000076408 UP000030765 UP000009046 UP000075886 UP000245037 UP000075903 UP000215335 UP000075881 UP000069272 UP000007062 UP000075884 UP000075900 UP000019118 UP000000673 UP000095300 UP000095301 UP000001070 UP000037069 UP000183832 UP000092443 UP000008792 UP000075901 UP000009192 UP000000803 UP000092553 UP000008711 UP000002282 UP000000304 UP000001292 UP000007801 UP000008744 UP000001819 UP000091820 UP000192221 UP000268350
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9ITK2
A0A2A4K4E7
A0A2W1BNK6
A0A194PYI7
A0A194QUG9
A0A212FMF8
+ More
A0A2A3EB67 A0A088A7F7 A0A195CTX8 A0A151JZ88 A0A158NFD5 A0A1B6CT84 A0A1B6DYS0 A0A1B6CEC5 A0A023F4B7 A0A3L8DGP3 A0A0C9Q4D2 A0A0L7LTD7 K7IW01 A0A069DZ97 J9JT06 A0A0M8ZRY9 A0A0J7L5W3 A0A1Y1NB36 T1I6X0 A0A151XFM2 A0A1Y1NB73 A0A2S2Q6F7 A0A1B6M7T8 A0A310SRE3 A0A2H8TF89 A0A2S2NP74 A0A1B6G7H6 A0A1W4WFI9 A0A1W4W3P0 A0A1W4WE96 A0A1W4W3P9 A0A139WL24 A0A2J7QTX9 V5I8T1 A0A182G9R9 A0A146M0F1 B0W7P2 A0A182GWR4 A0A1Q3FR00 A0A182JCV9 Q17DS4 A0A182XXX1 A0A084VQ76 E0W1U1 A0A182QC59 A0A2P8YI88 A0A182UV07 A0A232F9S6 A0A034W050 A0A182KH76 A0A182FIU1 W8AD07 A0A0A9YMX1 Q7PNJ1 F5HLY6 A0A0K8V7B1 A0A0A1WEG1 A0A182NV40 A0A182RMU1 N6TTC5 W5JS93 A0A1I8PPG6 A0A034W1Q1 A0A336LQI1 T1PC15 B4JN53 A0A0A1XKD8 Q6BCZ5 A0A0L0C1N1 A0A1J1IIF3 A0A1A9YQC3 A0A1L8D975 B4M1K1 A0A182SBW0 B4L229 X2JFT8 A0A0M4ELF5 B7Z155 B3NWA1 B4Q1X3 Q9Y115 B4R656 B4IEV4 B3N0B8 B4HA36 Q29FL5 Q6BCZ8 Q6BD00 Q6BCZ7 Q6BCZ1 A0A1A9WHR3 A0A1W4VWV5 A0A3B0J7N1
A0A2A3EB67 A0A088A7F7 A0A195CTX8 A0A151JZ88 A0A158NFD5 A0A1B6CT84 A0A1B6DYS0 A0A1B6CEC5 A0A023F4B7 A0A3L8DGP3 A0A0C9Q4D2 A0A0L7LTD7 K7IW01 A0A069DZ97 J9JT06 A0A0M8ZRY9 A0A0J7L5W3 A0A1Y1NB36 T1I6X0 A0A151XFM2 A0A1Y1NB73 A0A2S2Q6F7 A0A1B6M7T8 A0A310SRE3 A0A2H8TF89 A0A2S2NP74 A0A1B6G7H6 A0A1W4WFI9 A0A1W4W3P0 A0A1W4WE96 A0A1W4W3P9 A0A139WL24 A0A2J7QTX9 V5I8T1 A0A182G9R9 A0A146M0F1 B0W7P2 A0A182GWR4 A0A1Q3FR00 A0A182JCV9 Q17DS4 A0A182XXX1 A0A084VQ76 E0W1U1 A0A182QC59 A0A2P8YI88 A0A182UV07 A0A232F9S6 A0A034W050 A0A182KH76 A0A182FIU1 W8AD07 A0A0A9YMX1 Q7PNJ1 F5HLY6 A0A0K8V7B1 A0A0A1WEG1 A0A182NV40 A0A182RMU1 N6TTC5 W5JS93 A0A1I8PPG6 A0A034W1Q1 A0A336LQI1 T1PC15 B4JN53 A0A0A1XKD8 Q6BCZ5 A0A0L0C1N1 A0A1J1IIF3 A0A1A9YQC3 A0A1L8D975 B4M1K1 A0A182SBW0 B4L229 X2JFT8 A0A0M4ELF5 B7Z155 B3NWA1 B4Q1X3 Q9Y115 B4R656 B4IEV4 B3N0B8 B4HA36 Q29FL5 Q6BCZ8 Q6BD00 Q6BCZ7 Q6BCZ1 A0A1A9WHR3 A0A1W4VWV5 A0A3B0J7N1
Ontologies
Topology
Subcellular location
Membrane
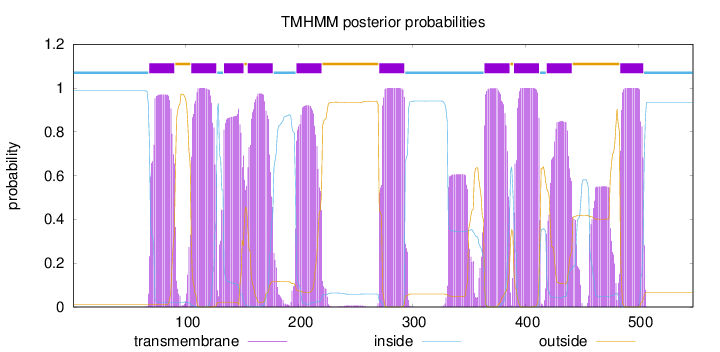
Length:
548
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
229.20011
Exp number, first 60 AAs:
0
Total prob of N-in:
0.98754
inside
1 - 67
TMhelix
68 - 90
outside
91 - 104
TMhelix
105 - 127
inside
128 - 133
TMhelix
134 - 151
outside
152 - 154
TMhelix
155 - 177
inside
178 - 197
TMhelix
198 - 220
outside
221 - 270
TMhelix
271 - 293
inside
294 - 363
TMhelix
364 - 386
outside
387 - 389
TMhelix
390 - 412
inside
413 - 418
TMhelix
419 - 441
outside
442 - 483
TMhelix
484 - 504
inside
505 - 548
Population Genetic Test Statistics
Pi
14.052458
Theta
17.67875
Tajima's D
-2.704928
CLR
0.265962
CSRT
0
Interpretation
Possibly Positive selection
