Gene
KWMTBOMO00205
Pre Gene Modal
BGIBMGA000584
Annotation
PREDICTED:_peptidoglycan-recognition_protein_LF-like_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.496
Sequence
CDS
ATGTATTCTGTTGTTAGTTTAAAAATTGCCAGAAATTATGGTTTTGTTTTAGGCCAAATACTCGGTCAGGAGCTGATATCATCTAAGAGCACACGCAAACTCCGCTGTAGTATCGCCGTGTTTGTTTGCTGGACGCTCATTGTCACATTAGGACTGGGCTTCTACATATCCCACTACGCGCTATCCAAGCTCACACGTCTCGATCTAGATATACACGAGCCCTGGTATCTTCGTCGAAGTGACTGGCAAGCGATGTCGCCATACTCAGTGGATTTCCTCGATTTACCCCTATCCTTCGTCATAGTTGGTCACAGCGCTACCAACTACTGTACTGAAAAGTACGAATGTATTAAGGAAATGTTGGATGTGCAGAAAAGCCATTTGCATCGCGGATGGCAAGATATCGGTCCCAATTTCCTCGTGTCTGGTAACGGCATCGTATTCGAAGGCAGAGGCGCTAATGTGTTCGGTGCAATGGCGATAGCTTGGAACCGCAGAAGCATACTAATCATGTTCCTCGGGGATTATACAAAGGATAAGACGACACCTGTACAGTTTGAACACTTAAATATCGTCTTAGATCAACTTGTAAAACAAGGAGTCTTGCGACCTGATTACACTTTGTATGGCCAATGCCAGGTAGCGTCGCTCACTATAAGTCCTGGACCTAACGTTATACAACTTCCACTGCCGCGCTTCATATGGAAAGTGTTGAAGAATACAAGTAGAGTGGAAAGATCGCTATGTGCTGCTGCTGTTTCAGCTTTAATCATATGTATAGCGTTGACACTTTACTTCACACTGTCAGCCAAGGCGACCACTGACGACGATGTTGCACCTCACCCATGGAACATCTCTAGAGATATGTGGCTGGCCAAGAAGTACAATGGGTCAGAAGCAATTACACAGTTTGTGCCGTTGCGACTGGTCATCGTCCAACACACGGTGAGCGCGAAATGCTCCAACTTTATCACGTGTGCGGCCGAAGTCCGGATCATCCAAACGTTTTATTTGGAAAAACAAAAATATGACATTCCTTACAATTTCATAATTGGCAACGACAATAGAGTTTATGAAGGACGCGGTTGGGGCATTATGGGAGCACATACTTTATCTTATAATAGCTGTTCGGTCGGTGTGGCTTTTATAGGTGATTACCGCGAGCTGGAAGCCACAGATCTTCAGATTAATAGAACACAGATGTTACTTGACGACGGCGTAAGAAGGGGTTTCCTACATCCCGATTACTATATTGTTGGTGCTTGTGATATCCGTGAAACAGTCAGTCCGGGAATCAACCTGTATAATGCCTTAAAAAAATTGAAGCATTTTGATCACAGCGGACGTTTCAAACACAAAACTTGTACGCAAATCTATGCAATGATCGAAAGTGAAAACTAA
Protein
MYSVVSLKIARNYGFVLGQILGQELISSKSTRKLRCSIAVFVCWTLIVTLGLGFYISHYALSKLTRLDLDIHEPWYLRRSDWQAMSPYSVDFLDLPLSFVIVGHSATNYCTEKYECIKEMLDVQKSHLHRGWQDIGPNFLVSGNGIVFEGRGANVFGAMAIAWNRRSILIMFLGDYTKDKTTPVQFEHLNIVLDQLVKQGVLRPDYTLYGQCQVASLTISPGPNVIQLPLPRFIWKVLKNTSRVERSLCAAAVSALIICIALTLYFTLSAKATTDDDVAPHPWNISRDMWLAKKYNGSEAITQFVPLRLVIVQHTVSAKCSNFITCAAEVRIIQTFYLEKQKYDIPYNFIIGNDNRVYEGRGWGIMGAHTLSYNSCSVGVAFIGDYRELEATDLQINRTQMLLDDGVRRGFLHPDYYIVGACDIRETVSPGINLYNALKKLKHFDHSGRFKHKTCTQIYAMIESEN
Summary
Uniprot
Pubmed
EMBL
KQ461108
KPJ09174.1
AGBW02007654
OWR54870.1
JTDY01006104
KOB66282.1
+ More
GDQN01007870 JAT83184.1 GDQN01004611 JAT86443.1 GDQN01010384 GDQN01002099 JAT80670.1 JAT88955.1 AGBW02007651 OWR54947.1 KQ459582 KPI98913.1 AEFK01161767 AEFK01161768 AEFK01161769 AEFK01161770 GALX01005044 JAB63422.1 AGTP01066417 RAZU01000055 RLQ77309.1 AANG04004008 KE663882 ERE90694.1 AABR07012289 JH001491 EGW10419.1 AABR07012273 AABR07012274 AB295597 BAK61786.2
GDQN01007870 JAT83184.1 GDQN01004611 JAT86443.1 GDQN01010384 GDQN01002099 JAT80670.1 JAT88955.1 AGBW02007651 OWR54947.1 KQ459582 KPI98913.1 AEFK01161767 AEFK01161768 AEFK01161769 AEFK01161770 GALX01005044 JAB63422.1 AGTP01066417 RAZU01000055 RLQ77309.1 AANG04004008 KE663882 ERE90694.1 AABR07012289 JH001491 EGW10419.1 AABR07012273 AABR07012274 AB295597 BAK61786.2
Proteomes
Pfam
PF01510 Amidase_2
Interpro
SUPFAM
SSF55846
SSF55846
Gene 3D
CDD
ProteinModelPortal
PDB
1SXR
E-value=5.3284e-28,
Score=310
Ontologies
GO
PANTHER
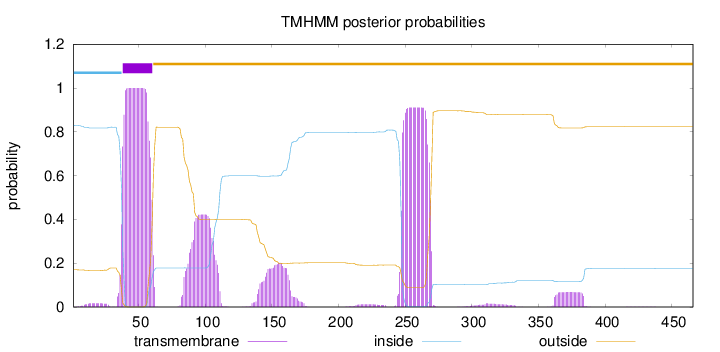
Topology
Length:
466
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
57.84489
Exp number, first 60 AAs:
22.28271
Total prob of N-in:
0.82946
POSSIBLE N-term signal
sequence
inside
1 - 37
TMhelix
38 - 60
outside
61 - 466
Population Genetic Test Statistics
Pi
0.289123
Theta
11.910197
Tajima's D
-2.2225
CLR
0.672519
CSRT
0.00469976501174941
Interpretation
Uncertain
