Gene
KWMTBOMO00201
Pre Gene Modal
BGIBMGA000628
Annotation
bestrophin_2_[Spodoptera_littoralis]
Full name
Bestrophin homolog
Location in the cell
PlasmaMembrane Reliability : 2.804
Sequence
CDS
ATGACTATATCGTACGCCGGTGAAGTACCAAACGGCAGCAGTTTTGGTTGTTTCTGGAGGATCCTCTGCAAATGGAGAGGCAGCGTCTACAAGCTGGTGTGGCGCGAGCTGCTAGCATACCTATTGCTTTACTATACAATAAACTTATTGTATCGCTTCGCACTCACAGAACAGCAACAAAGGGTGTTCGAGAAGGTTCGGCAGTATTTCGGAGCACAGAGCGAATCTATTCCGATGTCGTTCGTGCTCGGTTTCTACGTAAGTCTCGTCGTGAAGCGTTGGTGGGAACAGTACAAGCTCCTTCCGTGGCCTGATACGCTCGCGCTGTTCATCTCTGCAGGCATACCTGGAGCGGACGAGACGGGCCGATTAATGAGAAGAAATATTGTAAGATATGCTATCCTCGCCTACGTCATCACGCTACAGAGGGTGTCTCTTAGGGTAAAAAGAAGATTTCCAACATGGCAACACGTAGTTGACTCTGGTCTTATGTTGGAAAGCGAGAGAAAAGTTTTTGAAAAGATGGACGGCAAGAGTCCTATGTCCAAGTATTGGATGCCGTTAGTATGGGCGACGAATATAATAAACAGAGCCAGGAAAGAAGGCCTAATAACCAGTGACCACATAGTGCAAACGTTGTTGGTGGAGCTATCTGACATAAGACGAAGATTGGGGGCGCTCATTGGTTATGACACCGTTTGCGTACCACTCGTTTATACGCAGGTGGTAACTTTGTCCCTGTACACTTATTTCGTTGCGGCATTGATGGGCCGTCAACTGGTCCCTCCGGCGCCCGGAAGTACCTCCAAGTACGAACCTGACGTATACTTTCCGCTCTTCACAGCTTTACAGTTCTGCTTTTACGTGGGATGGTTGAAAGTAGCTGAAGTTTTAATTAATCCTTTCGGAGAAGATGACGATGACATCGAACTAAATTGGCTTATTGACAGGCACATAAAGGCAGCCTATATGATAGTTGACGAGATGCACGAAGAGCATCCTGAACTGCTGAAAGATCAATACTGGGAAGAGGTGGTCCCCAAAGATTTACCGTACACCGTAGCCTCAGAACATTACCGTCGCCATGAGCCCCCTTGCTCAGCTGACCACTACAAAGTAAAAGCAGAGGACGCTGTCTATGCTAACGTTCAAGCACCCAGAAAGAGTCATGATGAAACTTACGCTGATTACGAAAGTGTGGACACTCCCTTAGTTGAAAGGCGAAAAAATTGGTTCCAGCGACAAATATCGAGGATGGGCTCGGTACGGTCCGCTTCAACGGCGTATTCATCGGGAGGCCTCTTTGGACGAAATCGACACAATTCTGTTGTCTATTCGAGTCCTGAAGCAGGCCAGCCCGTGGCGCCACCTCCCCCGCACCACAAGATGTCTTTATATGAGCGACTTGTTGGTCGCAAATCGGGCAGAGGGCAGCATCGACAAAATTCAAGACACGGTGGCCAAAAAAGTAATGGATCCGCCGTTCCTATTACCCTGCGGAACAGACCTCGTATACCCACACCGGACGTGACGAAAGAGGTCATGGACCGTGAAAATCGTATCGCAATGGGTATGCAGAACATGGGTGTAATAATGGCTCACCAAGGCTATCAAAATGAAGTTCCCGTATTAGGTGCACTGGTCTTATCGCCTATACAGGAATTGGATAGTGGTTCCGTGAACAACACATTACACGCAGGGCAGCCTGGCACCACGGCTCTAGCGCAAGCAGTGCTGGCACCCGGTGGACTAACACCAATGCTCACTACTGCACCTGTTAACTTAACACCGATGGGTGTATCCCAGCTCACAACTATAGTGAGCTCAGCGCCATCGACGCCACGAGCTGAGCGTGGCCCAGCAGATGGCTCCGGTGGTTCACCGCAATCTCCAAGAGCGACCATTACAGAGCTCCCTCCATCGGATCGTGAAAGTAATCATAGCGGCACACCTCCAGATTTCGCGAGGAAACCAGGTTCTAAAAGAGGAGAGAGCAACGCGACTGACACAATTGAAGGAACTTCATTCGAACCAAGCGAAGAGCTCGTATCAGATGTTGAATGA
Protein
MTISYAGEVPNGSSFGCFWRILCKWRGSVYKLVWRELLAYLLLYYTINLLYRFALTEQQQRVFEKVRQYFGAQSESIPMSFVLGFYVSLVVKRWWEQYKLLPWPDTLALFISAGIPGADETGRLMRRNIVRYAILAYVITLQRVSLRVKRRFPTWQHVVDSGLMLESERKVFEKMDGKSPMSKYWMPLVWATNIINRARKEGLITSDHIVQTLLVELSDIRRRLGALIGYDTVCVPLVYTQVVTLSLYTYFVAALMGRQLVPPAPGSTSKYEPDVYFPLFTALQFCFYVGWLKVAEVLINPFGEDDDDIELNWLIDRHIKAAYMIVDEMHEEHPELLKDQYWEEVVPKDLPYTVASEHYRRHEPPCSADHYKVKAEDAVYANVQAPRKSHDETYADYESVDTPLVERRKNWFQRQISRMGSVRSASTAYSSGGLFGRNRHNSVVYSSPEAGQPVAPPPPHHKMSLYERLVGRKSGRGQHRQNSRHGGQKSNGSAVPITLRNRPRIPTPDVTKEVMDRENRIAMGMQNMGVIMAHQGYQNEVPVLGALVLSPIQELDSGSVNNTLHAGQPGTTALAQAVLAPGGLTPMLTTAPVNLTPMGVSQLTTIVSSAPSTPRAERGPADGSGGSPQSPRATITELPPSDRESNHSGTPPDFARKPGSKRGESNATDTIEGTSFEPSEELVSDVE
Summary
Description
Forms chloride channels.
Similarity
Belongs to the bestrophin family.
Uniprot
I6XZ71
A0A2A4JER2
A0A2A4JDB2
A0A2W1BJG2
A0A212FMA6
A0A194PTX5
+ More
A0A194QUH4 H9ITP8 A0A2H1VSW4 A0A139WKD3 A0A1Y1MQW5 A0A1L8DF52 A0A088A9S5 U5ETV9 A0A154NZ73 A0A0L7QZT4 A0A195CUG9 A0A0L0CPV1 A0A151IVZ2 A0A026W780 A0A158NQ03 A0A1B0GME3 A0A1B0G266 Q16KY4 A0A1A9XJI8 K7J614 A0A023EW33 A0A1Q3G4Z1 A0A1Q3G4Y9 A0A182FDZ8 A0A182WCU5 A0A182R0G2 A0A182NNJ4 B4N3X8 A0A182HXQ6 A0A1B6FHL1 A0A182YBS6 A0A1B6IYJ9 Q5TSK2 A0A182RKJ1 A0A0K8VPW4 A0A2M4A4M6 A0A182J9S7 A0A1B0A6L1 A0A084VXI9 W5J5D1 A0A2M3YZW2 A0A2M3YZU5 A0A1I8MK18 A0A2M4BE00 A0A2M4BE02 B3NGG8 Q9VRW4 B4HUT2 B4QJD0 B4PJN1 B4LI87 Q7YU35 A0A1W4V9D2 B3M9Z2 A0A182JP97 B4IXY4 A0A1B0AY46 A0A151XG23 A0A1B6KF69 A0A1A9UJ92 B4KVQ1 A0A146KT61 A0A1B6LGZ4 A0A195FND4 A0A195BI48 A0A1B6KR18 A0A182MLT6 A0A1B6LCH1 A0A182PSZ0 A0A182S870 A0A0P8YC90 A0A182LHZ9 A0A0J9RPT4 A0A1W4UXH2 A0A0Q5U493 A0A0Q9WJG1 A0A3B0KFP7 A0A182XNL5 A0A0R1DXQ5 B5DP29 A0A069DWF1 A0A0P4VMQ9 A0A1I8NQ24 A0A0K8UV58 A0A0K8WHV1 A0A023F3F5 J9JWQ3 A0A0V0G7D3 A0A0K8V2B3 A0A1B0G267 E0W1P6 A0A1J1IQX1
A0A194QUH4 H9ITP8 A0A2H1VSW4 A0A139WKD3 A0A1Y1MQW5 A0A1L8DF52 A0A088A9S5 U5ETV9 A0A154NZ73 A0A0L7QZT4 A0A195CUG9 A0A0L0CPV1 A0A151IVZ2 A0A026W780 A0A158NQ03 A0A1B0GME3 A0A1B0G266 Q16KY4 A0A1A9XJI8 K7J614 A0A023EW33 A0A1Q3G4Z1 A0A1Q3G4Y9 A0A182FDZ8 A0A182WCU5 A0A182R0G2 A0A182NNJ4 B4N3X8 A0A182HXQ6 A0A1B6FHL1 A0A182YBS6 A0A1B6IYJ9 Q5TSK2 A0A182RKJ1 A0A0K8VPW4 A0A2M4A4M6 A0A182J9S7 A0A1B0A6L1 A0A084VXI9 W5J5D1 A0A2M3YZW2 A0A2M3YZU5 A0A1I8MK18 A0A2M4BE00 A0A2M4BE02 B3NGG8 Q9VRW4 B4HUT2 B4QJD0 B4PJN1 B4LI87 Q7YU35 A0A1W4V9D2 B3M9Z2 A0A182JP97 B4IXY4 A0A1B0AY46 A0A151XG23 A0A1B6KF69 A0A1A9UJ92 B4KVQ1 A0A146KT61 A0A1B6LGZ4 A0A195FND4 A0A195BI48 A0A1B6KR18 A0A182MLT6 A0A1B6LCH1 A0A182PSZ0 A0A182S870 A0A0P8YC90 A0A182LHZ9 A0A0J9RPT4 A0A1W4UXH2 A0A0Q5U493 A0A0Q9WJG1 A0A3B0KFP7 A0A182XNL5 A0A0R1DXQ5 B5DP29 A0A069DWF1 A0A0P4VMQ9 A0A1I8NQ24 A0A0K8UV58 A0A0K8WHV1 A0A023F3F5 J9JWQ3 A0A0V0G7D3 A0A0K8V2B3 A0A1B0G267 E0W1P6 A0A1J1IQX1
Pubmed
23300744
28756777
22118469
26354079
19121390
18362917
+ More
19820115 28004739 26108605 24508170 30249741 21347285 17510324 20075255 24945155 17994087 25244985 12364791 14747013 17210077 24438588 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 18057021 26823975 20966253 15632085 26334808 27129103 25474469 20566863
19820115 28004739 26108605 24508170 30249741 21347285 17510324 20075255 24945155 17994087 25244985 12364791 14747013 17210077 24438588 20920257 23761445 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 17550304 18057021 26823975 20966253 15632085 26334808 27129103 25474469 20566863
EMBL
JQ968535
AFN57625.1
NWSH01001804
PCG70074.1
PCG70075.1
KZ150006
+ More
PZC75212.1 AGBW02007654 OWR54873.1 KQ459594 KPI96209.1 KQ461108 KPJ09178.1 BABH01006768 ODYU01004250 SOQ43923.1 KQ971331 KYB28301.1 GEZM01024027 JAV88049.1 GFDF01009069 JAV05015.1 GANO01002604 JAB57267.1 KQ434783 KZC04891.1 KQ414676 KOC64072.1 KQ977279 KYN04331.1 JRES01000089 KNC34222.1 KQ980880 KYN11995.1 KK107388 QOIP01000001 EZA51476.1 RLU26758.1 ADTU01022825 ADTU01022826 ADTU01022827 ADTU01022828 ADTU01022829 AJVK01011427 AJVK01011428 AJVK01011429 CCAG010019771 CH477932 CH477829 EAT34965.1 EAT35845.1 GAPW01000282 JAC13316.1 GFDL01000174 JAV34871.1 GFDL01000177 JAV34868.1 AXCN02001915 CH964095 EDW79333.1 APCN01005336 GECZ01020081 JAS49688.1 GECU01015713 JAS91993.1 AAAB01008905 EAL40404.3 GDHF01011402 JAI40912.1 GGFK01002351 MBW35672.1 ATLV01018075 ATLV01018076 KE525212 KFB42683.1 ADMH02002184 ETN57959.1 GGFM01001059 MBW21810.1 GGFM01001036 MBW21787.1 GGFJ01002135 MBW51276.1 GGFJ01002134 MBW51275.1 CH954178 EDV51134.1 AE014296 BT099579 AAF50668.2 ACU43541.1 CH480817 EDW50703.1 CM000363 CM002912 EDX09439.1 KMY97910.1 CM000159 EDW93630.1 CH940647 EDW69654.1 KRF84497.1 BT010012 AAQ22481.1 CH902618 EDV41212.1 KPU79004.1 CH916366 EDV96505.1 JXJN01005545 JXJN01005546 KQ982182 KYQ59230.1 GEBQ01029912 JAT10065.1 CH933809 EDW18425.2 GDHC01018945 JAP99683.1 GEBQ01017001 JAT22976.1 KQ981382 KYN42075.1 KQ976464 KYM84500.1 GEBQ01026102 JAT13875.1 AXCM01012182 GEBQ01018579 JAT21398.1 KPU79003.1 KMY97911.1 KQS43675.1 KRF84498.1 OUUW01000009 SPP85149.1 KRK01278.1 CH379067 EDY73280.2 GBGD01000649 JAC88240.1 GDKW01002984 JAI53611.1 GDHF01021730 JAI30584.1 GDHF01001665 JAI50649.1 GBBI01002864 JAC15848.1 ABLF02030258 ABLF02030261 ABLF02062784 GECL01002090 JAP04034.1 GDHF01019381 GDHF01017132 JAI32933.1 JAI35182.1 DS235873 EEB19628.1 CVRI01000058 CRL02637.1
PZC75212.1 AGBW02007654 OWR54873.1 KQ459594 KPI96209.1 KQ461108 KPJ09178.1 BABH01006768 ODYU01004250 SOQ43923.1 KQ971331 KYB28301.1 GEZM01024027 JAV88049.1 GFDF01009069 JAV05015.1 GANO01002604 JAB57267.1 KQ434783 KZC04891.1 KQ414676 KOC64072.1 KQ977279 KYN04331.1 JRES01000089 KNC34222.1 KQ980880 KYN11995.1 KK107388 QOIP01000001 EZA51476.1 RLU26758.1 ADTU01022825 ADTU01022826 ADTU01022827 ADTU01022828 ADTU01022829 AJVK01011427 AJVK01011428 AJVK01011429 CCAG010019771 CH477932 CH477829 EAT34965.1 EAT35845.1 GAPW01000282 JAC13316.1 GFDL01000174 JAV34871.1 GFDL01000177 JAV34868.1 AXCN02001915 CH964095 EDW79333.1 APCN01005336 GECZ01020081 JAS49688.1 GECU01015713 JAS91993.1 AAAB01008905 EAL40404.3 GDHF01011402 JAI40912.1 GGFK01002351 MBW35672.1 ATLV01018075 ATLV01018076 KE525212 KFB42683.1 ADMH02002184 ETN57959.1 GGFM01001059 MBW21810.1 GGFM01001036 MBW21787.1 GGFJ01002135 MBW51276.1 GGFJ01002134 MBW51275.1 CH954178 EDV51134.1 AE014296 BT099579 AAF50668.2 ACU43541.1 CH480817 EDW50703.1 CM000363 CM002912 EDX09439.1 KMY97910.1 CM000159 EDW93630.1 CH940647 EDW69654.1 KRF84497.1 BT010012 AAQ22481.1 CH902618 EDV41212.1 KPU79004.1 CH916366 EDV96505.1 JXJN01005545 JXJN01005546 KQ982182 KYQ59230.1 GEBQ01029912 JAT10065.1 CH933809 EDW18425.2 GDHC01018945 JAP99683.1 GEBQ01017001 JAT22976.1 KQ981382 KYN42075.1 KQ976464 KYM84500.1 GEBQ01026102 JAT13875.1 AXCM01012182 GEBQ01018579 JAT21398.1 KPU79003.1 KMY97911.1 KQS43675.1 KRF84498.1 OUUW01000009 SPP85149.1 KRK01278.1 CH379067 EDY73280.2 GBGD01000649 JAC88240.1 GDKW01002984 JAI53611.1 GDHF01021730 JAI30584.1 GDHF01001665 JAI50649.1 GBBI01002864 JAC15848.1 ABLF02030258 ABLF02030261 ABLF02062784 GECL01002090 JAP04034.1 GDHF01019381 GDHF01017132 JAI32933.1 JAI35182.1 DS235873 EEB19628.1 CVRI01000058 CRL02637.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000005204
UP000007266
+ More
UP000005203 UP000076502 UP000053825 UP000078542 UP000037069 UP000078492 UP000053097 UP000279307 UP000005205 UP000092462 UP000092444 UP000008820 UP000092443 UP000002358 UP000069272 UP000075920 UP000075886 UP000075884 UP000007798 UP000075840 UP000076408 UP000007062 UP000075900 UP000075880 UP000092445 UP000030765 UP000000673 UP000095301 UP000008711 UP000000803 UP000001292 UP000000304 UP000002282 UP000008792 UP000192221 UP000007801 UP000075881 UP000001070 UP000092460 UP000075809 UP000078200 UP000009192 UP000078541 UP000078540 UP000075883 UP000075885 UP000075901 UP000075882 UP000268350 UP000076407 UP000001819 UP000095300 UP000007819 UP000009046 UP000183832
UP000005203 UP000076502 UP000053825 UP000078542 UP000037069 UP000078492 UP000053097 UP000279307 UP000005205 UP000092462 UP000092444 UP000008820 UP000092443 UP000002358 UP000069272 UP000075920 UP000075886 UP000075884 UP000007798 UP000075840 UP000076408 UP000007062 UP000075900 UP000075880 UP000092445 UP000030765 UP000000673 UP000095301 UP000008711 UP000000803 UP000001292 UP000000304 UP000002282 UP000008792 UP000192221 UP000007801 UP000075881 UP000001070 UP000092460 UP000075809 UP000078200 UP000009192 UP000078541 UP000078540 UP000075883 UP000075885 UP000075901 UP000075882 UP000268350 UP000076407 UP000001819 UP000095300 UP000007819 UP000009046 UP000183832
Pfam
PF01062 Bestrophin
ProteinModelPortal
I6XZ71
A0A2A4JER2
A0A2A4JDB2
A0A2W1BJG2
A0A212FMA6
A0A194PTX5
+ More
A0A194QUH4 H9ITP8 A0A2H1VSW4 A0A139WKD3 A0A1Y1MQW5 A0A1L8DF52 A0A088A9S5 U5ETV9 A0A154NZ73 A0A0L7QZT4 A0A195CUG9 A0A0L0CPV1 A0A151IVZ2 A0A026W780 A0A158NQ03 A0A1B0GME3 A0A1B0G266 Q16KY4 A0A1A9XJI8 K7J614 A0A023EW33 A0A1Q3G4Z1 A0A1Q3G4Y9 A0A182FDZ8 A0A182WCU5 A0A182R0G2 A0A182NNJ4 B4N3X8 A0A182HXQ6 A0A1B6FHL1 A0A182YBS6 A0A1B6IYJ9 Q5TSK2 A0A182RKJ1 A0A0K8VPW4 A0A2M4A4M6 A0A182J9S7 A0A1B0A6L1 A0A084VXI9 W5J5D1 A0A2M3YZW2 A0A2M3YZU5 A0A1I8MK18 A0A2M4BE00 A0A2M4BE02 B3NGG8 Q9VRW4 B4HUT2 B4QJD0 B4PJN1 B4LI87 Q7YU35 A0A1W4V9D2 B3M9Z2 A0A182JP97 B4IXY4 A0A1B0AY46 A0A151XG23 A0A1B6KF69 A0A1A9UJ92 B4KVQ1 A0A146KT61 A0A1B6LGZ4 A0A195FND4 A0A195BI48 A0A1B6KR18 A0A182MLT6 A0A1B6LCH1 A0A182PSZ0 A0A182S870 A0A0P8YC90 A0A182LHZ9 A0A0J9RPT4 A0A1W4UXH2 A0A0Q5U493 A0A0Q9WJG1 A0A3B0KFP7 A0A182XNL5 A0A0R1DXQ5 B5DP29 A0A069DWF1 A0A0P4VMQ9 A0A1I8NQ24 A0A0K8UV58 A0A0K8WHV1 A0A023F3F5 J9JWQ3 A0A0V0G7D3 A0A0K8V2B3 A0A1B0G267 E0W1P6 A0A1J1IQX1
A0A194QUH4 H9ITP8 A0A2H1VSW4 A0A139WKD3 A0A1Y1MQW5 A0A1L8DF52 A0A088A9S5 U5ETV9 A0A154NZ73 A0A0L7QZT4 A0A195CUG9 A0A0L0CPV1 A0A151IVZ2 A0A026W780 A0A158NQ03 A0A1B0GME3 A0A1B0G266 Q16KY4 A0A1A9XJI8 K7J614 A0A023EW33 A0A1Q3G4Z1 A0A1Q3G4Y9 A0A182FDZ8 A0A182WCU5 A0A182R0G2 A0A182NNJ4 B4N3X8 A0A182HXQ6 A0A1B6FHL1 A0A182YBS6 A0A1B6IYJ9 Q5TSK2 A0A182RKJ1 A0A0K8VPW4 A0A2M4A4M6 A0A182J9S7 A0A1B0A6L1 A0A084VXI9 W5J5D1 A0A2M3YZW2 A0A2M3YZU5 A0A1I8MK18 A0A2M4BE00 A0A2M4BE02 B3NGG8 Q9VRW4 B4HUT2 B4QJD0 B4PJN1 B4LI87 Q7YU35 A0A1W4V9D2 B3M9Z2 A0A182JP97 B4IXY4 A0A1B0AY46 A0A151XG23 A0A1B6KF69 A0A1A9UJ92 B4KVQ1 A0A146KT61 A0A1B6LGZ4 A0A195FND4 A0A195BI48 A0A1B6KR18 A0A182MLT6 A0A1B6LCH1 A0A182PSZ0 A0A182S870 A0A0P8YC90 A0A182LHZ9 A0A0J9RPT4 A0A1W4UXH2 A0A0Q5U493 A0A0Q9WJG1 A0A3B0KFP7 A0A182XNL5 A0A0R1DXQ5 B5DP29 A0A069DWF1 A0A0P4VMQ9 A0A1I8NQ24 A0A0K8UV58 A0A0K8WHV1 A0A023F3F5 J9JWQ3 A0A0V0G7D3 A0A0K8V2B3 A0A1B0G267 E0W1P6 A0A1J1IQX1
PDB
6N23
E-value=6.48456e-96,
Score=898
Ontologies
PANTHER
Topology
Subcellular location
Cell membrane
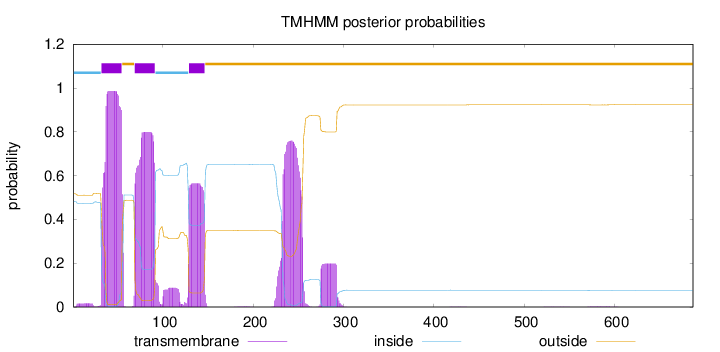
Length:
687
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
71.8834700000002
Exp number, first 60 AAs:
21.32043
Total prob of N-in:
0.48224
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 54
outside
55 - 68
TMhelix
69 - 91
inside
92 - 128
TMhelix
129 - 146
outside
147 - 687
Population Genetic Test Statistics
Pi
15.29601
Theta
17.944571
Tajima's D
-1.19496
CLR
0.071496
CSRT
0.102844857757112
Interpretation
Possibly Positive selection
