Gene
KWMTBOMO00199
Pre Gene Modal
BGIBMGA000587
Annotation
PREDICTED:_discoidin_domain-containing_receptor_2-like_isoform_X1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.402 Nuclear Reliability : 1.102
Sequence
CDS
ATGGAGAGCGGCCTGATTGCCGACAAGGACGTCACTGCTAGTTCTTGTTATAACGATGGCAACGTCGCGCCGCAGAACGGAAGGCTAAATCAAGACATCAAAGGAGGTGCATGGTGTCCTAGATCCCAAATCACGACTGAATCCACAGAGTGGCTAGAAATTGACCTTCACACTGTACACGTAATCACCGGTGCAGGCACCCAGGGTCGTTTCGGTAACGGCCAAGGTCAAGAGTACGCGGAGGCCTACGTACTTGAATATTGGCGCCCTAAACTTGGGAAATGGGTACGATACAGGAGCTTAGACGGCCAAGAGAAACTGACCGGTAATACTAACACATACTTGGAAAAGAAGAATCATCTAGAGCCGCCGATCTGGGCGTCCAAAGTTCGTTTCATTCCTTACAGTTCGCATAGACGGACTGTTTGTATGAGAGTTGAATTGTATGGATGCTATTGGAGCGTGGGAGTGGTTTCGTATTCGATGCCTAAAGGCGATAGACGTAGTAATGGAATCGAACTCACGGACATGATCTACGACGGTCAGTGGGGCGAGGAGCTCCGCGGGGGCTTGGGCCAACTGGTGGACGGTCAGTTCGGAAGCGACGACATCCGAGAAGCCTCTAAGAACTCCGCCTGGGTCGGCTGGAGAAACGACTCGAGACCAGATCCCCCGGTCATCATATTCGAATTCGACAAAGTCCGCGAGTTCAGCGCTGTACATATATTTTGTAATAATAAGTTCATGAGGGATGTACTAGTGTTCTCTGAAGCAATTATATCATTTTCGATCGGCGGTCGTCACTTTCAAGACGAACCTATCCACTATACATACGTTCAAGACAAGATCTTCGAGAACGCTCGTAACGTGAGCATTAAACTACACCATAGAATAGGGAAGTGGGTGCGGATCGAACTTCAGTTTGCCGCGAGGTGGATACTTGTTAGCGAGATCGTTTTTGATTCAGACGTAGCACAAGGCAATTACACACCGGAAAGTATGAAGCCACCGGTCAGTAAGGATAAAACGAAAGTGATTTCAAGTAAAGAGGTACCTATTTCGACGGCCCATCAGGAAGATCCGCTGTACGTCGCGATTGTCGTGGGAGTGTTGACCGCTCTCGTCATACTGTTGGCCGTTGCGATATTCCTTATAATACACCGACACAAACATAGGAAGTGTTTCGCTTCTCCGCTTGCGAAGTCGGCTGTCTCGCAGAAACGGGCCGTTGAGAGCTACAGTGGGTGCGGTACGTCTATGCTACCCGAGAACAAAATGATCTTAGACTGCGGTCTCGATGTCAAGTCAGACGAGTACCAGGAGCCCTATCAGGCTCTCAAATGCGCTCCCTACTTTAGTTACAGCACGGTCCTGCTCGAGATGAAAGATTTTAAGGACTCCAACACCGCGCTCTCTGACAGTTCGAACTACGATTATGCGGTACCAGAATTGAGTTCGGCTCCGTTGTTAACGAAGCGGATGGGAGAAACGCTTTCCCAGCAAGCTACCGAGCTTGTTGGCGAGCCGATGGACACGTTACCTGACCTCGGAACGAGACGATCCCTCAATCCTTCCGTACGATCCAAACAAAGCACGAGATCGCCGAGTCAACAAGAAATATTGTTGGATCTGAAGAGACGCCTCGAAACTACTAACGTTATTGAGTTCCCGAGGCACAGACTCCGGATGATCTCTAAGTTAGCCGAGGGCGCCTTCGGAACCGTTTACGTAGCGGAAGCGGACGGTGTGCCTGAATATAACGGAGCCATTACGTCGGAGAAGAGATTAGTGGCCGTCAAATTCTTATGTCACGACGCCACGCTCAAGGAGAGGTATATCATGGAGGAGTTCGAACGCGACGTCCGCATATTGGCCGCGTTGTCCTCGCCTCACCTCGCGAGGGTATTGGGCGCGTGTCGCTCGCCGCCCCTCGCCGTGGTGCTGGAGTACCTTGAGCTCGGGGACCTGTGTGCGTTCCTGCGGGCTGCTATTCCGCCGACGGCAGCCACGCTCCTGCACATTGCCACGCAGATAGCCGCTGGAATGCAATACCTCGAATCGCTCAATTTCGTTCACAGGGACTTAGCAGCTAGGAATTGTCTGATCGGGAAGGGTTATCAAATTAAAATAAGCGATTTCGGCACCGACAACGAGGCGTACACTTGCGACTATTATAAAGTGGACGGTCGCGTACCGCTGCCTCTGCGCTGGGCGGCCTGGGAGTCGGTGCTCCGGGGAAAGTACACGACCAAGAGCGACGTGTGGGCGTTCGCGGTCACGCTGCACGAGATATACTCGCTCTGCCGACGGCGCCCCTACGAGCACCTAACCGACTGCGAGGTTTTGGATAATCTGTCGCACTTACAAGCGGACGACGGTATGTTCGAGTGCGTCGACCGCGCCCCCGGCTGTCCCCGGGACCTCTACGAGCTGATGCTGGAATGCTGGCGGAGAGACGAACTGGACCGGCCCACGTTCGGTGAGATCCATCTCTTCCTGCAGCGCAAGAGTCTCAACTACGCGACCACGTGA
Protein
MESGLIADKDVTASSCYNDGNVAPQNGRLNQDIKGGAWCPRSQITTESTEWLEIDLHTVHVITGAGTQGRFGNGQGQEYAEAYVLEYWRPKLGKWVRYRSLDGQEKLTGNTNTYLEKKNHLEPPIWASKVRFIPYSSHRRTVCMRVELYGCYWSVGVVSYSMPKGDRRSNGIELTDMIYDGQWGEELRGGLGQLVDGQFGSDDIREASKNSAWVGWRNDSRPDPPVIIFEFDKVREFSAVHIFCNNKFMRDVLVFSEAIISFSIGGRHFQDEPIHYTYVQDKIFENARNVSIKLHHRIGKWVRIELQFAARWILVSEIVFDSDVAQGNYTPESMKPPVSKDKTKVISSKEVPISTAHQEDPLYVAIVVGVLTALVILLAVAIFLIIHRHKHRKCFASPLAKSAVSQKRAVESYSGCGTSMLPENKMILDCGLDVKSDEYQEPYQALKCAPYFSYSTVLLEMKDFKDSNTALSDSSNYDYAVPELSSAPLLTKRMGETLSQQATELVGEPMDTLPDLGTRRSLNPSVRSKQSTRSPSQQEILLDLKRRLETTNVIEFPRHRLRMISKLAEGAFGTVYVAEADGVPEYNGAITSEKRLVAVKFLCHDATLKERYIMEEFERDVRILAALSSPHLARVLGACRSPPLAVVLEYLELGDLCAFLRAAIPPTAATLLHIATQIAAGMQYLESLNFVHRDLAARNCLIGKGYQIKISDFGTDNEAYTCDYYKVDGRVPLPLRWAAWESVLRGKYTTKSDVWAFAVTLHEIYSLCRRRPYEHLTDCEVLDNLSHLQADDGMFECVDRAPGCPRDLYELMLECWRRDELDRPTFGEIHLFLQRKSLNYATT
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
H9ITK7
A0A2H1VWV3
A0A2W1BNK7
A0A194QW37
A0A212FMA3
A0A2A4JAE7
+ More
A0A2A4JBL3 A0A194PSB9 A0A195FPJ0 A0A3L8DQC9 A0A146LSE5 A0A067R7Z8 A0A0C9RLC0 A0A1W4WM45 A0A1B6LKK5 D6WFT6 A0A232FAP8 A0A1Y1LZ85 A0A158NFP8 J9JU75 A0A2H8TKL8 A0A2S2P6A0 A0A2S2QVE2 A0A2A3EKL6 E0W014 A0A151WPD5 A0A0U3U674 A0A087ZZC4 T1I6J0 T1J7A6 A0A1B6DPL3 A0A226EK82 A0A310S6X0 A0A1B6I1Y9
A0A2A4JBL3 A0A194PSB9 A0A195FPJ0 A0A3L8DQC9 A0A146LSE5 A0A067R7Z8 A0A0C9RLC0 A0A1W4WM45 A0A1B6LKK5 D6WFT6 A0A232FAP8 A0A1Y1LZ85 A0A158NFP8 J9JU75 A0A2H8TKL8 A0A2S2P6A0 A0A2S2QVE2 A0A2A3EKL6 E0W014 A0A151WPD5 A0A0U3U674 A0A087ZZC4 T1I6J0 T1J7A6 A0A1B6DPL3 A0A226EK82 A0A310S6X0 A0A1B6I1Y9
Pubmed
EMBL
BABH01006764
BABH01006765
BABH01006766
ODYU01004929
SOQ45310.1
KZ150006
+ More
PZC75214.1 KQ461108 KPJ09180.1 AGBW02007654 OWR54875.1 NWSH01002247 PCG68819.1 PCG68820.1 KQ459594 KPI96207.1 KQ981340 KYN42505.1 QOIP01000005 RLU22645.1 GDHC01009279 JAQ09350.1 KK852638 KDR19671.1 GBYB01009070 JAG78837.1 GEBQ01015779 JAT24198.1 KQ971319 EEZ99592.2 NNAY01000610 OXU27407.1 GEZM01046289 JAV77355.1 ADTU01014467 ADTU01014468 ADTU01014469 ADTU01014470 ADTU01014471 ADTU01014472 ADTU01014473 ADTU01014474 ADTU01014475 ADTU01014476 ABLF02040549 GFXV01002880 MBW14685.1 GGMR01012402 MBY25021.1 GGMS01012515 MBY81718.1 KZ288230 PBC31571.1 DS235854 EEB18970.1 KQ982877 KYQ49690.1 KT355578 ALV82497.1 ACPB03008763 JH431916 GEDC01009674 JAS27624.1 LNIX01000003 OXA57161.1 KQ767065 OAD53524.1 GECU01026831 JAS80875.1
PZC75214.1 KQ461108 KPJ09180.1 AGBW02007654 OWR54875.1 NWSH01002247 PCG68819.1 PCG68820.1 KQ459594 KPI96207.1 KQ981340 KYN42505.1 QOIP01000005 RLU22645.1 GDHC01009279 JAQ09350.1 KK852638 KDR19671.1 GBYB01009070 JAG78837.1 GEBQ01015779 JAT24198.1 KQ971319 EEZ99592.2 NNAY01000610 OXU27407.1 GEZM01046289 JAV77355.1 ADTU01014467 ADTU01014468 ADTU01014469 ADTU01014470 ADTU01014471 ADTU01014472 ADTU01014473 ADTU01014474 ADTU01014475 ADTU01014476 ABLF02040549 GFXV01002880 MBW14685.1 GGMR01012402 MBY25021.1 GGMS01012515 MBY81718.1 KZ288230 PBC31571.1 DS235854 EEB18970.1 KQ982877 KYQ49690.1 KT355578 ALV82497.1 ACPB03008763 JH431916 GEDC01009674 JAS27624.1 LNIX01000003 OXA57161.1 KQ767065 OAD53524.1 GECU01026831 JAS80875.1
Proteomes
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9ITK7
A0A2H1VWV3
A0A2W1BNK7
A0A194QW37
A0A212FMA3
A0A2A4JAE7
+ More
A0A2A4JBL3 A0A194PSB9 A0A195FPJ0 A0A3L8DQC9 A0A146LSE5 A0A067R7Z8 A0A0C9RLC0 A0A1W4WM45 A0A1B6LKK5 D6WFT6 A0A232FAP8 A0A1Y1LZ85 A0A158NFP8 J9JU75 A0A2H8TKL8 A0A2S2P6A0 A0A2S2QVE2 A0A2A3EKL6 E0W014 A0A151WPD5 A0A0U3U674 A0A087ZZC4 T1I6J0 T1J7A6 A0A1B6DPL3 A0A226EK82 A0A310S6X0 A0A1B6I1Y9
A0A2A4JBL3 A0A194PSB9 A0A195FPJ0 A0A3L8DQC9 A0A146LSE5 A0A067R7Z8 A0A0C9RLC0 A0A1W4WM45 A0A1B6LKK5 D6WFT6 A0A232FAP8 A0A1Y1LZ85 A0A158NFP8 J9JU75 A0A2H8TKL8 A0A2S2P6A0 A0A2S2QVE2 A0A2A3EKL6 E0W014 A0A151WPD5 A0A0U3U674 A0A087ZZC4 T1I6J0 T1J7A6 A0A1B6DPL3 A0A226EK82 A0A310S6X0 A0A1B6I1Y9
PDB
5BVW
E-value=8.23698e-71,
Score=682
Ontologies
GO
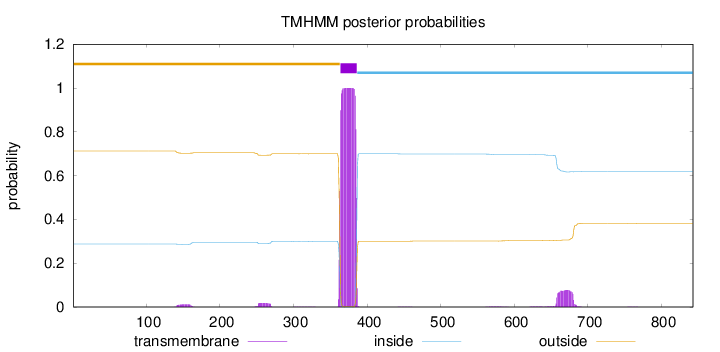
Topology
Length:
843
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.40534
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.28767
outside
1 - 363
TMhelix
364 - 386
inside
387 - 843
Population Genetic Test Statistics
Pi
17.863335
Theta
17.629928
Tajima's D
0.060399
CLR
0.0916
CSRT
0.396030198490075
Interpretation
Uncertain
