Pre Gene Modal
BGIBMGA000626
Annotation
PREDICTED:_exosome_component_10_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.423 Nuclear Reliability : 1.373
Sequence
CDS
ATGAACTCCGCGCTTAATCCAGATGCACCTGAGTTTTATCCACACCTGGCGGCAGTCACACAGGAAGGATATAGAAGTGTTAACAAGACAATAAAGTCGTCAAATCAACTGCCGATGAGCTCTTCGTATGATTTATTTAAAACATATCCAGAATTTAATGAAATTTCAAATCAGATCAGCCAAAATGTTGCTCAACAGTCAAACGAACTGATTGGCACTGAACAACCGGCCGTGAAGCTGAAGTGTGAGGATTTCACAAGCAACATAGAGAAACTTGCTGATGTAAACGACAGCTTATTGGACAGGATTAATATTAGCATTGATACAGTGAGCGGTGAGAACTTACCCAGTGAGGCATTCGCGAGCGGTCTCAATGATAATCCTTCTCATAACTCATCGCCTTGGAATAAGCCTGTCATTTCTGAAAAGAAGATGGGATCAACTATATTCATCGGAGCTAAAAACATTCCGCGTCCACAGCTTACTTTCAAGGATACAATTGATAATTCTGAGAATTTGTGGGTGCCCAAAATTTCAGACAAACCAAACAACATCAAGCCTTTGGCTTTAAACATTTTGTATACCGAGAAGGGCGAAGCTGTTGGATATGAGCATCCATACCAAATGGAGTTGGACATGTATAACCCTCCGTCGCAGTTCATCGATCCCGATCCGGAGCCTCCCATGTTTCCACCCCCTCTGGAGGAAACGAAATTCACTTACATAGATATAGAATCGAAGCTAGACGAGTTGGTTGAACACCTTATGGCCGTCGAACAAATTGCTGTAGATGTAGAACATCATTCCTATAGGACCTATCAGGGCATAACGTGCCTCATTCAAATATCAACTGATGAAGGAGGAGACTTTATCATTGACGCTCTCGCTGTAAGGGAACACATACATAAACTGAATGTCGTCTTCACTGACCCCAAGAAACTAAAGGTGTTCCACGGTGCCGATAGCGATGTTCTTTGGCTTCAACGTGATTTCGGCGTGTATTTGGTGGGTCTTTTCGACACATACCACGCAGCCAAGTCACTAGGTCTGCCCGCGCTTTCGCTCAAATTTCTTTTGATGAAGTATTGCGGCGTTGACACCGATAAAACGTGA
Protein
MNSALNPDAPEFYPHLAAVTQEGYRSVNKTIKSSNQLPMSSSYDLFKTYPEFNEISNQISQNVAQQSNELIGTEQPAVKLKCEDFTSNIEKLADVNDSLLDRINISIDTVSGENLPSEAFASGLNDNPSHNSSPWNKPVISEKKMGSTIFIGAKNIPRPQLTFKDTIDNSENLWVPKISDKPNNIKPLALNILYTEKGEAVGYEHPYQMELDMYNPPSQFIDPDPEPPMFPPPLEETKFTYIDIESKLDELVEHLMAVEQIAVDVEHHSYRTYQGITCLIQISTDEGGDFIIDALAVREHIHKLNVVFTDPKKLKVFHGADSDVLWLQRDFGVYLVGLFDTYHAAKSLGLPALSLKFLLMKYCGVDTDKT
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9ITP6
A0A2A4JBP7
A0A2H1VWX8
A0A2A4J9R3
A0A2W1BRB1
A0A212EMN4
+ More
A0A0L7L075 S4PRA6 A0A194QUN7 A0A194PYH6 A0A0J7NIP3 A0A0P5V8E6 A0A182PEM0 A0A3L8DR98 A0A026WCC8 K7J5C7 A0A0L7RCV5 E9IEY4 A0A195DCF1 A0A182TRL6 A0A182K798 A0A182LFN7 A0A182XLZ3 A0A0P6D6F6 A0A0N8CBZ7 A0A182HTH4 A0A195F2Z0 F5HK87 Q7QBT6 A0A0P5WQZ6 A0A0P5TW28 A0A158P390 A0A1J1IKT1 A0A0P5UVH9 A0A0N8CL65 A0A0N8C7L1 A0A0P5TW33 A0A0P5IP63 A0A0P4YR15 A0A088ARS5 A0A182VEF2 A0A0P5IK07 A0A1Z5LCK5 E2AJL2 A0A182JDM0 A0A0N8CDV7 A0A0P5B4E1 A0A0P4YM19 F4WM69 A0A0P5E570 A0A195BD00 A0A0P5V1U6 A0A0P5W350 A0A0P4ZB77 A0A0P5MPF3 A0A0P5ECA4 A0A0P5V8E5 A0A0P5V845 A0A151WFD4 A0A0P5YFZ2 A0A0M8ZMM5 A0A0P5G515 A0A0P5UAS4 A0A151IGE3 A0A182W2M2 A0A154PA47 A0A182QEF5 A0A182NG92 A0A1Y1KTQ2 A0A2M4BBY9 A0A2M4BBW4 A0A2M4BCJ6 A0A182MCX6 A0A0P5TLJ6 A0A182XVE5 E9GKT7 A0A2A3E2G0 A0A3B0K1C5 A0A0P4YSW2 W5JI40 A0A023FN87 A0A2M3Z3W2 A0A2P8YHT9 A0A182R756 B4PSN8 B4NJ59 A0A0P6BGD3 A0A1W4W0P5 A0A224YSY7 Q29AI2 A0A1S4FNZ8 A0A2M4A807 A0A2M4A816 A0A2M4A8Q1 A0A0L0CBS3 A0A1I8M6P5 A0A182FQ03 A0A1I8PA12
A0A0L7L075 S4PRA6 A0A194QUN7 A0A194PYH6 A0A0J7NIP3 A0A0P5V8E6 A0A182PEM0 A0A3L8DR98 A0A026WCC8 K7J5C7 A0A0L7RCV5 E9IEY4 A0A195DCF1 A0A182TRL6 A0A182K798 A0A182LFN7 A0A182XLZ3 A0A0P6D6F6 A0A0N8CBZ7 A0A182HTH4 A0A195F2Z0 F5HK87 Q7QBT6 A0A0P5WQZ6 A0A0P5TW28 A0A158P390 A0A1J1IKT1 A0A0P5UVH9 A0A0N8CL65 A0A0N8C7L1 A0A0P5TW33 A0A0P5IP63 A0A0P4YR15 A0A088ARS5 A0A182VEF2 A0A0P5IK07 A0A1Z5LCK5 E2AJL2 A0A182JDM0 A0A0N8CDV7 A0A0P5B4E1 A0A0P4YM19 F4WM69 A0A0P5E570 A0A195BD00 A0A0P5V1U6 A0A0P5W350 A0A0P4ZB77 A0A0P5MPF3 A0A0P5ECA4 A0A0P5V8E5 A0A0P5V845 A0A151WFD4 A0A0P5YFZ2 A0A0M8ZMM5 A0A0P5G515 A0A0P5UAS4 A0A151IGE3 A0A182W2M2 A0A154PA47 A0A182QEF5 A0A182NG92 A0A1Y1KTQ2 A0A2M4BBY9 A0A2M4BBW4 A0A2M4BCJ6 A0A182MCX6 A0A0P5TLJ6 A0A182XVE5 E9GKT7 A0A2A3E2G0 A0A3B0K1C5 A0A0P4YSW2 W5JI40 A0A023FN87 A0A2M3Z3W2 A0A2P8YHT9 A0A182R756 B4PSN8 B4NJ59 A0A0P6BGD3 A0A1W4W0P5 A0A224YSY7 Q29AI2 A0A1S4FNZ8 A0A2M4A807 A0A2M4A816 A0A2M4A8Q1 A0A0L0CBS3 A0A1I8M6P5 A0A182FQ03 A0A1I8PA12
Pubmed
EMBL
BABH01006761
BABH01006762
BABH01006763
NWSH01002247
PCG68823.1
ODYU01004929
+ More
SOQ45308.1 PCG68825.1 KZ150006 PZC75216.1 AGBW02013811 OWR42755.1 JTDY01003871 KOB68897.1 GAIX01000970 JAA91590.1 KQ461108 KPJ09182.1 KQ459594 KPI96205.1 LBMM01004468 KMQ92385.1 GDIP01102924 JAM00791.1 QOIP01000005 RLU22965.1 KK107293 EZA53331.1 AAZX01008278 AAZX01019459 AAZX01019480 KQ414614 KOC68822.1 GL762728 EFZ20873.1 KQ980989 KYN10566.1 GDIQ01090032 JAN04705.1 GDIQ01094683 JAL57043.1 APCN01000528 KQ981855 KYN34751.1 AAAB01008859 EGK96928.1 EAA07463.5 GDIP01082835 JAM20880.1 GDIP01120926 JAL82788.1 ADTU01001310 CVRI01000054 CRL00784.1 GDIP01108235 JAL95479.1 GDIP01120927 JAL82787.1 GDIQ01106971 JAL44755.1 GDIP01120925 JAL82789.1 GDIQ01212384 JAK39341.1 GDIP01225866 JAI97535.1 GDIQ01212383 JAK39342.1 GFJQ02001838 JAW05132.1 GL440027 EFN66407.1 GDIP01141391 JAL62323.1 GDIP01189827 JAJ33575.1 GDIP01225867 JAI97534.1 GL888217 EGI64713.1 GDIP01146384 JAJ77018.1 KQ976522 KYM82087.1 GDIP01108234 GDIP01084631 LRGB01002190 JAL95480.1 KZS08544.1 GDIP01105728 JAL97986.1 GDIP01216570 JAJ06832.1 GDIQ01176190 JAK75535.1 GDIP01143535 JAJ79867.1 GDIP01102925 JAM00790.1 GDIP01105729 JAL97985.1 KQ983219 KYQ46537.1 GDIP01059289 JAM44426.1 KQ438551 KOX67177.1 GDIQ01246184 JAK05541.1 GDIP01115677 JAL88037.1 KQ977726 KYN00296.1 KQ434856 KZC08697.1 AXCN02001628 GEZM01073849 JAV64782.1 GGFJ01001399 MBW50540.1 GGFJ01001398 MBW50539.1 GGFJ01001397 MBW50538.1 AXCM01000235 GDIP01126038 JAL77676.1 GL732550 EFX79910.1 KZ288427 PBC25885.1 OUUW01000013 SPP88034.1 GDIP01223508 JAI99893.1 ADMH02001428 ETN62570.1 GBBK01002147 JAC22335.1 GGFM01002460 MBW23211.1 PYGN01000583 PSN43819.1 CM000160 EDW97534.1 CH964272 EDW83852.1 GDIP01015024 JAM88691.1 GFPF01007713 MAA18859.1 CM000070 EAL27368.1 GGFK01003612 MBW36933.1 GGFK01003622 MBW36943.1 GGFK01003862 MBW37183.1 JRES01000641 KNC29695.1
SOQ45308.1 PCG68825.1 KZ150006 PZC75216.1 AGBW02013811 OWR42755.1 JTDY01003871 KOB68897.1 GAIX01000970 JAA91590.1 KQ461108 KPJ09182.1 KQ459594 KPI96205.1 LBMM01004468 KMQ92385.1 GDIP01102924 JAM00791.1 QOIP01000005 RLU22965.1 KK107293 EZA53331.1 AAZX01008278 AAZX01019459 AAZX01019480 KQ414614 KOC68822.1 GL762728 EFZ20873.1 KQ980989 KYN10566.1 GDIQ01090032 JAN04705.1 GDIQ01094683 JAL57043.1 APCN01000528 KQ981855 KYN34751.1 AAAB01008859 EGK96928.1 EAA07463.5 GDIP01082835 JAM20880.1 GDIP01120926 JAL82788.1 ADTU01001310 CVRI01000054 CRL00784.1 GDIP01108235 JAL95479.1 GDIP01120927 JAL82787.1 GDIQ01106971 JAL44755.1 GDIP01120925 JAL82789.1 GDIQ01212384 JAK39341.1 GDIP01225866 JAI97535.1 GDIQ01212383 JAK39342.1 GFJQ02001838 JAW05132.1 GL440027 EFN66407.1 GDIP01141391 JAL62323.1 GDIP01189827 JAJ33575.1 GDIP01225867 JAI97534.1 GL888217 EGI64713.1 GDIP01146384 JAJ77018.1 KQ976522 KYM82087.1 GDIP01108234 GDIP01084631 LRGB01002190 JAL95480.1 KZS08544.1 GDIP01105728 JAL97986.1 GDIP01216570 JAJ06832.1 GDIQ01176190 JAK75535.1 GDIP01143535 JAJ79867.1 GDIP01102925 JAM00790.1 GDIP01105729 JAL97985.1 KQ983219 KYQ46537.1 GDIP01059289 JAM44426.1 KQ438551 KOX67177.1 GDIQ01246184 JAK05541.1 GDIP01115677 JAL88037.1 KQ977726 KYN00296.1 KQ434856 KZC08697.1 AXCN02001628 GEZM01073849 JAV64782.1 GGFJ01001399 MBW50540.1 GGFJ01001398 MBW50539.1 GGFJ01001397 MBW50538.1 AXCM01000235 GDIP01126038 JAL77676.1 GL732550 EFX79910.1 KZ288427 PBC25885.1 OUUW01000013 SPP88034.1 GDIP01223508 JAI99893.1 ADMH02001428 ETN62570.1 GBBK01002147 JAC22335.1 GGFM01002460 MBW23211.1 PYGN01000583 PSN43819.1 CM000160 EDW97534.1 CH964272 EDW83852.1 GDIP01015024 JAM88691.1 GFPF01007713 MAA18859.1 CM000070 EAL27368.1 GGFK01003612 MBW36933.1 GGFK01003622 MBW36943.1 GGFK01003862 MBW37183.1 JRES01000641 KNC29695.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000036403 UP000075885 UP000279307 UP000053097 UP000002358 UP000053825 UP000078492 UP000075902 UP000075881 UP000075882 UP000076407 UP000075840 UP000078541 UP000007062 UP000005205 UP000183832 UP000005203 UP000075903 UP000000311 UP000075880 UP000007755 UP000078540 UP000076858 UP000075809 UP000053105 UP000078542 UP000075920 UP000076502 UP000075886 UP000075884 UP000075883 UP000076408 UP000000305 UP000242457 UP000268350 UP000000673 UP000245037 UP000075900 UP000002282 UP000007798 UP000192221 UP000001819 UP000037069 UP000095301 UP000069272 UP000095300
UP000036403 UP000075885 UP000279307 UP000053097 UP000002358 UP000053825 UP000078492 UP000075902 UP000075881 UP000075882 UP000076407 UP000075840 UP000078541 UP000007062 UP000005205 UP000183832 UP000005203 UP000075903 UP000000311 UP000075880 UP000007755 UP000078540 UP000076858 UP000075809 UP000053105 UP000078542 UP000075920 UP000076502 UP000075886 UP000075884 UP000075883 UP000076408 UP000000305 UP000242457 UP000268350 UP000000673 UP000245037 UP000075900 UP000002282 UP000007798 UP000192221 UP000001819 UP000037069 UP000095301 UP000069272 UP000095300
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9ITP6
A0A2A4JBP7
A0A2H1VWX8
A0A2A4J9R3
A0A2W1BRB1
A0A212EMN4
+ More
A0A0L7L075 S4PRA6 A0A194QUN7 A0A194PYH6 A0A0J7NIP3 A0A0P5V8E6 A0A182PEM0 A0A3L8DR98 A0A026WCC8 K7J5C7 A0A0L7RCV5 E9IEY4 A0A195DCF1 A0A182TRL6 A0A182K798 A0A182LFN7 A0A182XLZ3 A0A0P6D6F6 A0A0N8CBZ7 A0A182HTH4 A0A195F2Z0 F5HK87 Q7QBT6 A0A0P5WQZ6 A0A0P5TW28 A0A158P390 A0A1J1IKT1 A0A0P5UVH9 A0A0N8CL65 A0A0N8C7L1 A0A0P5TW33 A0A0P5IP63 A0A0P4YR15 A0A088ARS5 A0A182VEF2 A0A0P5IK07 A0A1Z5LCK5 E2AJL2 A0A182JDM0 A0A0N8CDV7 A0A0P5B4E1 A0A0P4YM19 F4WM69 A0A0P5E570 A0A195BD00 A0A0P5V1U6 A0A0P5W350 A0A0P4ZB77 A0A0P5MPF3 A0A0P5ECA4 A0A0P5V8E5 A0A0P5V845 A0A151WFD4 A0A0P5YFZ2 A0A0M8ZMM5 A0A0P5G515 A0A0P5UAS4 A0A151IGE3 A0A182W2M2 A0A154PA47 A0A182QEF5 A0A182NG92 A0A1Y1KTQ2 A0A2M4BBY9 A0A2M4BBW4 A0A2M4BCJ6 A0A182MCX6 A0A0P5TLJ6 A0A182XVE5 E9GKT7 A0A2A3E2G0 A0A3B0K1C5 A0A0P4YSW2 W5JI40 A0A023FN87 A0A2M3Z3W2 A0A2P8YHT9 A0A182R756 B4PSN8 B4NJ59 A0A0P6BGD3 A0A1W4W0P5 A0A224YSY7 Q29AI2 A0A1S4FNZ8 A0A2M4A807 A0A2M4A816 A0A2M4A8Q1 A0A0L0CBS3 A0A1I8M6P5 A0A182FQ03 A0A1I8PA12
A0A0L7L075 S4PRA6 A0A194QUN7 A0A194PYH6 A0A0J7NIP3 A0A0P5V8E6 A0A182PEM0 A0A3L8DR98 A0A026WCC8 K7J5C7 A0A0L7RCV5 E9IEY4 A0A195DCF1 A0A182TRL6 A0A182K798 A0A182LFN7 A0A182XLZ3 A0A0P6D6F6 A0A0N8CBZ7 A0A182HTH4 A0A195F2Z0 F5HK87 Q7QBT6 A0A0P5WQZ6 A0A0P5TW28 A0A158P390 A0A1J1IKT1 A0A0P5UVH9 A0A0N8CL65 A0A0N8C7L1 A0A0P5TW33 A0A0P5IP63 A0A0P4YR15 A0A088ARS5 A0A182VEF2 A0A0P5IK07 A0A1Z5LCK5 E2AJL2 A0A182JDM0 A0A0N8CDV7 A0A0P5B4E1 A0A0P4YM19 F4WM69 A0A0P5E570 A0A195BD00 A0A0P5V1U6 A0A0P5W350 A0A0P4ZB77 A0A0P5MPF3 A0A0P5ECA4 A0A0P5V8E5 A0A0P5V845 A0A151WFD4 A0A0P5YFZ2 A0A0M8ZMM5 A0A0P5G515 A0A0P5UAS4 A0A151IGE3 A0A182W2M2 A0A154PA47 A0A182QEF5 A0A182NG92 A0A1Y1KTQ2 A0A2M4BBY9 A0A2M4BBW4 A0A2M4BCJ6 A0A182MCX6 A0A0P5TLJ6 A0A182XVE5 E9GKT7 A0A2A3E2G0 A0A3B0K1C5 A0A0P4YSW2 W5JI40 A0A023FN87 A0A2M3Z3W2 A0A2P8YHT9 A0A182R756 B4PSN8 B4NJ59 A0A0P6BGD3 A0A1W4W0P5 A0A224YSY7 Q29AI2 A0A1S4FNZ8 A0A2M4A807 A0A2M4A816 A0A2M4A8Q1 A0A0L0CBS3 A0A1I8M6P5 A0A182FQ03 A0A1I8PA12
PDB
6D6R
E-value=5.88916e-49,
Score=490
Ontologies
GO
Topology
Subcellular location
Secreted
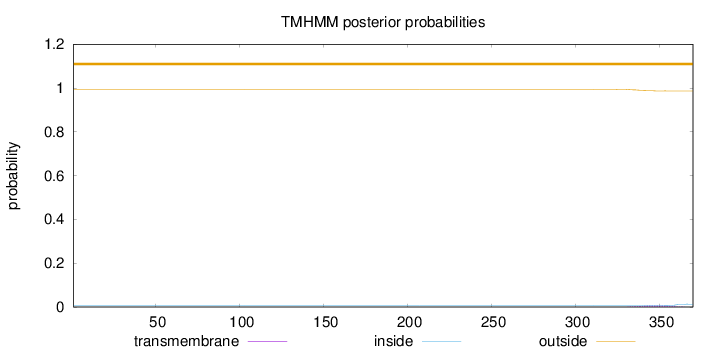
Length:
370
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.17261
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00585
outside
1 - 370
Population Genetic Test Statistics
Pi
0.763926
Theta
4.669905
Tajima's D
-2.51867
CLR
1.759402
CSRT
9.99950002499875e-05
Interpretation
Uncertain
