Gene
KWMTBOMO00192
Pre Gene Modal
BGIBMGA000590
Annotation
pleiotrophin-like_protein_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.453 Nuclear Reliability : 1.836
Sequence
CDS
ATGGAGCTCAAGTACTGGTGGTGGATGATGGCAGGACTGGCCCTCTTATCGGTAGCGGTAGTTGCCGCAGATGGCGAGGTGTGGGAGGAGAATGACCATGAAGTTCTCATTCGAAGCGCAAGAGGCGCAAAAAATAGAGAGGCATGTCGATATGTGCGCGGTGCTTGGAGCGAATGTGACTCCAAAACGAATATCCGCTCAAGGAAGTTGACTCTGAAAAAGGGCGATCCGGCTAACTGTGAAGTTGTTAAAACCATCCAGAAAAAGTGCAAGAGAACTTGCCGCTATGAAAAGTCATCTTGGAGCGAATGCAGCATCAATGGGGAGATGTCCAGGACCGATAAGCTAAAGTCCAACAGCGATTCGACTTGCGACCAAAGTAGGCGCAAAACTCGGAAATGTAACAAGAATAAGCAAGTCAAGCTTGCTAAAGATAAGGGTCGCAGAAATCGCCAGTAA
Protein
MELKYWWWMMAGLALLSVAVVAADGEVWEENDHEVLIRSARGAKNREACRYVRGAWSECDSKTNIRSRKLTLKKGDPANCEVVKTIQKKCKRTCRYEKSSWSECSINGEMSRTDKLKSNSDSTCDQSRRKTRKCNKNKQVKLAKDKGRRNRQ
Summary
Uniprot
Q8T100
A0A2W1BJJ7
A0A2A4JAV1
A0A2A4JAF9
A0A2H1WA31
I4DMR7
+ More
I4DNC7 A0A194PSB4 A0A1E1WLI9 A0A212EMT4 A0A194QW43 A0A1Y1MTG2 A0A146MBW7 A0A1W4XHJ5 R4V1F0 A0A1W4XSL1 D6WG44 A0A139WM48 A0A2J7R458 J3JWR8 A0A146M9S5 A0A0K8T454 U5EQG6 A0A1B6M5B8 A0A1B6JH17 A0A2K8JSB9 A0A1B0AUB2 A0A1B0FRJ0 A0A0L0BYX0 A0A1A9Z695 T1PHZ8 B4KXY4 A0A1A9XD31 A0A1I8N3P5 Q29ES1 B3M8Y0 A0A1W4W0L0 A0A3B0JXZ3 A0A1S4EFN5 B4PC43 B3NEN1 B4HVE0 B4QEY3 Q9W0S5 A0A0C9RB15 B4MN89 Q9Y0V9 A0A0Q9WBV4 B4MGR1 A0A034VZX4 B4IYK2 W8BK54 A0A1Q3FZU8 E2A0A5 T1I2V2 A0A023F222 A0A0M3QVT7 A0A069DP60 A0A1B6C3U0 A0A224XMF4 A0A1L8DP00 A0A2A3E5W0 V9IC14 A0A310S7M6 A0A088A554 T1H5T7 A0A0N0BIJ2 A0A151J0D3 F4WJC6 A0A195BVG0 A0A195FL12 A0A023EHP5 A0A0J7L581 A0A1S4EFZ1 A0A0C9RZW9 A0A154P8W8 A0A195CCF4 Q17PM5 A0A182HWQ5 A0A182NGY4 A0A182KE51 A0A182XMU8 Q7Q6T7 E0VZD6 A0A026W2X9 E2B646 A0A182PHP6 A0A182Y1C0 A0A1L8DPB3 K7J652 A0A1B0AUB7 A0A1A9XD37 A0A1B0FKU3
I4DNC7 A0A194PSB4 A0A1E1WLI9 A0A212EMT4 A0A194QW43 A0A1Y1MTG2 A0A146MBW7 A0A1W4XHJ5 R4V1F0 A0A1W4XSL1 D6WG44 A0A139WM48 A0A2J7R458 J3JWR8 A0A146M9S5 A0A0K8T454 U5EQG6 A0A1B6M5B8 A0A1B6JH17 A0A2K8JSB9 A0A1B0AUB2 A0A1B0FRJ0 A0A0L0BYX0 A0A1A9Z695 T1PHZ8 B4KXY4 A0A1A9XD31 A0A1I8N3P5 Q29ES1 B3M8Y0 A0A1W4W0L0 A0A3B0JXZ3 A0A1S4EFN5 B4PC43 B3NEN1 B4HVE0 B4QEY3 Q9W0S5 A0A0C9RB15 B4MN89 Q9Y0V9 A0A0Q9WBV4 B4MGR1 A0A034VZX4 B4IYK2 W8BK54 A0A1Q3FZU8 E2A0A5 T1I2V2 A0A023F222 A0A0M3QVT7 A0A069DP60 A0A1B6C3U0 A0A224XMF4 A0A1L8DP00 A0A2A3E5W0 V9IC14 A0A310S7M6 A0A088A554 T1H5T7 A0A0N0BIJ2 A0A151J0D3 F4WJC6 A0A195BVG0 A0A195FL12 A0A023EHP5 A0A0J7L581 A0A1S4EFZ1 A0A0C9RZW9 A0A154P8W8 A0A195CCF4 Q17PM5 A0A182HWQ5 A0A182NGY4 A0A182KE51 A0A182XMU8 Q7Q6T7 E0VZD6 A0A026W2X9 E2B646 A0A182PHP6 A0A182Y1C0 A0A1L8DPB3 K7J652 A0A1B0AUB7 A0A1A9XD37 A0A1B0FKU3
Pubmed
12715162
28756777
22651552
26354079
22118469
28004739
+ More
26823975 18362917 19820115 22516182 26108605 17994087 25315136 15632085 23185243 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 24495485 20798317 25474469 26334808 21719571 24945155 17510324 12364791 20566863 24508170 30249741 25244985 20075255
26823975 18362917 19820115 22516182 26108605 17994087 25315136 15632085 23185243 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25348373 24495485 20798317 25474469 26334808 21719571 24945155 17510324 12364791 20566863 24508170 30249741 25244985 20075255
EMBL
AB079868
AB090308
BAB85199.1
KZ150006
PZC75219.1
NWSH01002247
+ More
PCG68828.1 PCG68829.1 ODYU01007290 SOQ49941.1 AK402585 BAM19207.1 AK402850 BAM19417.1 KQ459594 KPI96202.1 GDQN01003333 JAT87721.1 AGBW02013811 OWR42751.1 KQ461108 KPJ09185.1 GEZM01021515 GEZM01021514 JAV88964.1 GDHC01002579 JAQ16050.1 KC571968 AGM32467.1 KQ971319 EFA00526.2 KYB28915.1 NEVH01007815 PNF35619.1 BT127686 AEE62648.1 GDHC01002121 JAQ16508.1 GBRD01005509 JAG60312.1 GANO01003273 JAB56598.1 GEBQ01008851 JAT31126.1 GECU01009238 JAS98468.1 KY031179 ATU82930.1 JXJN01003584 CCAG010004862 CCAG010004863 CCAG010004864 CCAG010004865 CCAG010004866 JRES01001236 KNC24434.1 KA648309 AFP62938.1 CH933809 EDW17656.1 CH379070 EAL29989.2 KRT08564.1 CH902618 EDV38924.1 OUUW01000012 SPP86924.1 CM000159 EDW92698.1 CH954178 EDV50026.1 CH480817 EDW49905.1 CM000362 CM002912 EDX07010.1 KMY96617.1 AE014296 AY071689 AAF47368.2 AAL49311.1 AAS64923.1 AGB93892.1 GBYB01013684 JAG83451.1 CH963847 EDW73645.1 AF149800 AAD37797.2 CH940682 KRF77616.1 EDW57127.2 KRF77617.1 GAKP01011527 JAC47425.1 CH916366 EDV95512.1 GAMC01007453 GAMC01007452 JAB99102.1 GFDL01001995 JAV33050.1 GL435569 EFN73108.1 ACPB03017904 ACPB03017905 GBBI01003319 JAC15393.1 CP012525 ALC42973.1 GBGD01003249 JAC85640.1 GEDC01029329 GEDC01014583 JAS07969.1 JAS22715.1 GFTR01002771 JAW13655.1 GFDF01005881 JAV08203.1 KZ288364 PBC26874.1 JR039271 AEY58610.1 KQ777311 OAD52139.1 KQ435729 KOX77622.1 KQ980636 KYN14949.1 GL888182 EGI65704.1 KQ976401 KYM92567.1 KQ981490 KYN41036.1 GAPW01004695 JAC08903.1 LBMM01000545 KMQ98092.1 GBYB01013686 JAG83453.1 KQ434846 KZC08355.1 KQ978023 KYM97911.1 CH477190 EAT48727.1 APCN01001523 AAAB01008960 EAA11318.3 DS235851 EEB18742.1 KK107455 QOIP01000010 EZA50435.1 RLU17665.1 GL445914 EFN88835.1 GFDF01005870 JAV08214.1 JXJN01003603 JXJN01003604 CCAG010000748 CCAG010000749
PCG68828.1 PCG68829.1 ODYU01007290 SOQ49941.1 AK402585 BAM19207.1 AK402850 BAM19417.1 KQ459594 KPI96202.1 GDQN01003333 JAT87721.1 AGBW02013811 OWR42751.1 KQ461108 KPJ09185.1 GEZM01021515 GEZM01021514 JAV88964.1 GDHC01002579 JAQ16050.1 KC571968 AGM32467.1 KQ971319 EFA00526.2 KYB28915.1 NEVH01007815 PNF35619.1 BT127686 AEE62648.1 GDHC01002121 JAQ16508.1 GBRD01005509 JAG60312.1 GANO01003273 JAB56598.1 GEBQ01008851 JAT31126.1 GECU01009238 JAS98468.1 KY031179 ATU82930.1 JXJN01003584 CCAG010004862 CCAG010004863 CCAG010004864 CCAG010004865 CCAG010004866 JRES01001236 KNC24434.1 KA648309 AFP62938.1 CH933809 EDW17656.1 CH379070 EAL29989.2 KRT08564.1 CH902618 EDV38924.1 OUUW01000012 SPP86924.1 CM000159 EDW92698.1 CH954178 EDV50026.1 CH480817 EDW49905.1 CM000362 CM002912 EDX07010.1 KMY96617.1 AE014296 AY071689 AAF47368.2 AAL49311.1 AAS64923.1 AGB93892.1 GBYB01013684 JAG83451.1 CH963847 EDW73645.1 AF149800 AAD37797.2 CH940682 KRF77616.1 EDW57127.2 KRF77617.1 GAKP01011527 JAC47425.1 CH916366 EDV95512.1 GAMC01007453 GAMC01007452 JAB99102.1 GFDL01001995 JAV33050.1 GL435569 EFN73108.1 ACPB03017904 ACPB03017905 GBBI01003319 JAC15393.1 CP012525 ALC42973.1 GBGD01003249 JAC85640.1 GEDC01029329 GEDC01014583 JAS07969.1 JAS22715.1 GFTR01002771 JAW13655.1 GFDF01005881 JAV08203.1 KZ288364 PBC26874.1 JR039271 AEY58610.1 KQ777311 OAD52139.1 KQ435729 KOX77622.1 KQ980636 KYN14949.1 GL888182 EGI65704.1 KQ976401 KYM92567.1 KQ981490 KYN41036.1 GAPW01004695 JAC08903.1 LBMM01000545 KMQ98092.1 GBYB01013686 JAG83453.1 KQ434846 KZC08355.1 KQ978023 KYM97911.1 CH477190 EAT48727.1 APCN01001523 AAAB01008960 EAA11318.3 DS235851 EEB18742.1 KK107455 QOIP01000010 EZA50435.1 RLU17665.1 GL445914 EFN88835.1 GFDF01005870 JAV08214.1 JXJN01003603 JXJN01003604 CCAG010000748 CCAG010000749
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000192223
UP000007266
+ More
UP000235965 UP000092460 UP000092444 UP000037069 UP000092445 UP000009192 UP000092443 UP000095301 UP000001819 UP000007801 UP000192221 UP000268350 UP000079169 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000007798 UP000008792 UP000001070 UP000000311 UP000015103 UP000092553 UP000242457 UP000005203 UP000015102 UP000053105 UP000078492 UP000007755 UP000078540 UP000078541 UP000036403 UP000076502 UP000078542 UP000008820 UP000075840 UP000075884 UP000075881 UP000076407 UP000007062 UP000009046 UP000053097 UP000279307 UP000008237 UP000075885 UP000076408 UP000002358
UP000235965 UP000092460 UP000092444 UP000037069 UP000092445 UP000009192 UP000092443 UP000095301 UP000001819 UP000007801 UP000192221 UP000268350 UP000079169 UP000002282 UP000008711 UP000001292 UP000000304 UP000000803 UP000007798 UP000008792 UP000001070 UP000000311 UP000015103 UP000092553 UP000242457 UP000005203 UP000015102 UP000053105 UP000078492 UP000007755 UP000078540 UP000078541 UP000036403 UP000076502 UP000078542 UP000008820 UP000075840 UP000075884 UP000075881 UP000076407 UP000007062 UP000009046 UP000053097 UP000279307 UP000008237 UP000075885 UP000076408 UP000002358
Pfam
PF01091 PTN_MK_C
SUPFAM
SSF57288
SSF57288
Gene 3D
ProteinModelPortal
Q8T100
A0A2W1BJJ7
A0A2A4JAV1
A0A2A4JAF9
A0A2H1WA31
I4DMR7
+ More
I4DNC7 A0A194PSB4 A0A1E1WLI9 A0A212EMT4 A0A194QW43 A0A1Y1MTG2 A0A146MBW7 A0A1W4XHJ5 R4V1F0 A0A1W4XSL1 D6WG44 A0A139WM48 A0A2J7R458 J3JWR8 A0A146M9S5 A0A0K8T454 U5EQG6 A0A1B6M5B8 A0A1B6JH17 A0A2K8JSB9 A0A1B0AUB2 A0A1B0FRJ0 A0A0L0BYX0 A0A1A9Z695 T1PHZ8 B4KXY4 A0A1A9XD31 A0A1I8N3P5 Q29ES1 B3M8Y0 A0A1W4W0L0 A0A3B0JXZ3 A0A1S4EFN5 B4PC43 B3NEN1 B4HVE0 B4QEY3 Q9W0S5 A0A0C9RB15 B4MN89 Q9Y0V9 A0A0Q9WBV4 B4MGR1 A0A034VZX4 B4IYK2 W8BK54 A0A1Q3FZU8 E2A0A5 T1I2V2 A0A023F222 A0A0M3QVT7 A0A069DP60 A0A1B6C3U0 A0A224XMF4 A0A1L8DP00 A0A2A3E5W0 V9IC14 A0A310S7M6 A0A088A554 T1H5T7 A0A0N0BIJ2 A0A151J0D3 F4WJC6 A0A195BVG0 A0A195FL12 A0A023EHP5 A0A0J7L581 A0A1S4EFZ1 A0A0C9RZW9 A0A154P8W8 A0A195CCF4 Q17PM5 A0A182HWQ5 A0A182NGY4 A0A182KE51 A0A182XMU8 Q7Q6T7 E0VZD6 A0A026W2X9 E2B646 A0A182PHP6 A0A182Y1C0 A0A1L8DPB3 K7J652 A0A1B0AUB7 A0A1A9XD37 A0A1B0FKU3
I4DNC7 A0A194PSB4 A0A1E1WLI9 A0A212EMT4 A0A194QW43 A0A1Y1MTG2 A0A146MBW7 A0A1W4XHJ5 R4V1F0 A0A1W4XSL1 D6WG44 A0A139WM48 A0A2J7R458 J3JWR8 A0A146M9S5 A0A0K8T454 U5EQG6 A0A1B6M5B8 A0A1B6JH17 A0A2K8JSB9 A0A1B0AUB2 A0A1B0FRJ0 A0A0L0BYX0 A0A1A9Z695 T1PHZ8 B4KXY4 A0A1A9XD31 A0A1I8N3P5 Q29ES1 B3M8Y0 A0A1W4W0L0 A0A3B0JXZ3 A0A1S4EFN5 B4PC43 B3NEN1 B4HVE0 B4QEY3 Q9W0S5 A0A0C9RB15 B4MN89 Q9Y0V9 A0A0Q9WBV4 B4MGR1 A0A034VZX4 B4IYK2 W8BK54 A0A1Q3FZU8 E2A0A5 T1I2V2 A0A023F222 A0A0M3QVT7 A0A069DP60 A0A1B6C3U0 A0A224XMF4 A0A1L8DP00 A0A2A3E5W0 V9IC14 A0A310S7M6 A0A088A554 T1H5T7 A0A0N0BIJ2 A0A151J0D3 F4WJC6 A0A195BVG0 A0A195FL12 A0A023EHP5 A0A0J7L581 A0A1S4EFZ1 A0A0C9RZW9 A0A154P8W8 A0A195CCF4 Q17PM5 A0A182HWQ5 A0A182NGY4 A0A182KE51 A0A182XMU8 Q7Q6T7 E0VZD6 A0A026W2X9 E2B646 A0A182PHP6 A0A182Y1C0 A0A1L8DPB3 K7J652 A0A1B0AUB7 A0A1A9XD37 A0A1B0FKU3
Ontologies
GO
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.992104
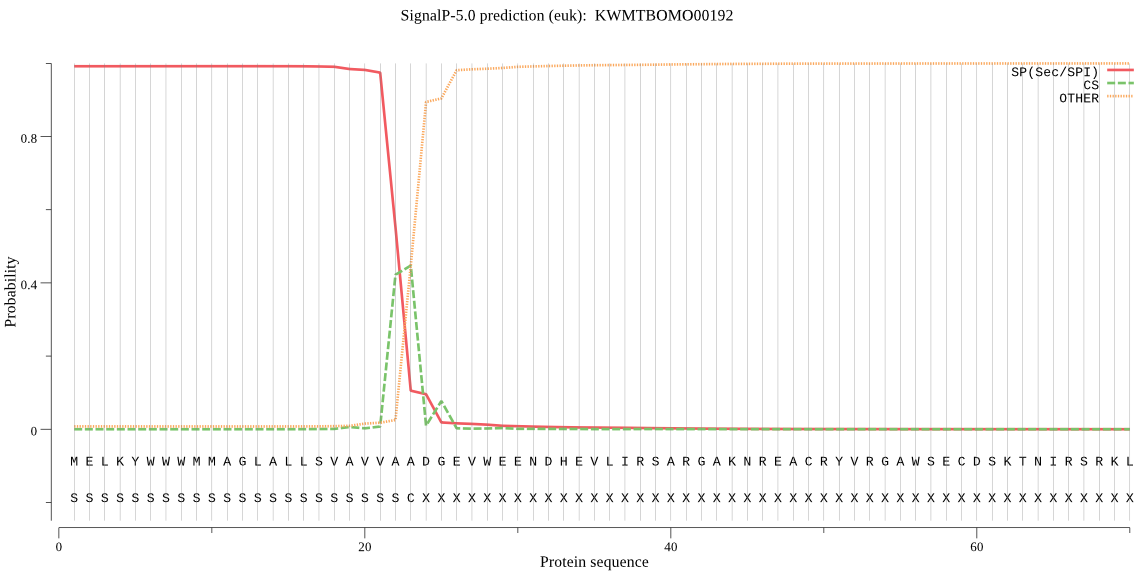
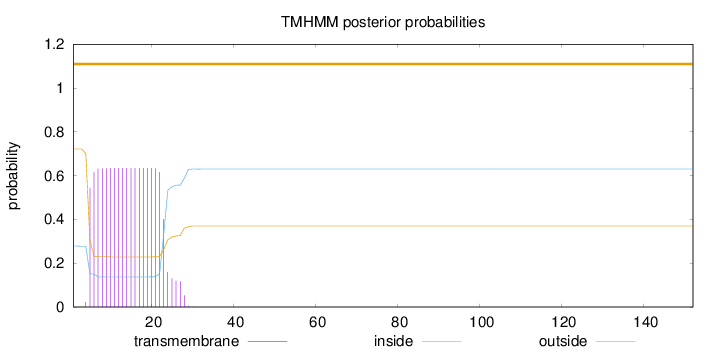
Length:
152
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.2973
Exp number, first 60 AAs:
12.2973
Total prob of N-in:
0.27844
POSSIBLE N-term signal
sequence
outside
1 - 152
Population Genetic Test Statistics
Pi
0.40259
Theta
4.599709
Tajima's D
-2.498756
CLR
1.061489
CSRT
0.000299985000749962
Interpretation
Possibly Positive selection
