Gene
KWMTBOMO00187
Pre Gene Modal
BGIBMGA000593
Annotation
PREDICTED:_hikaru_genki_isoform_X1_[Bombyx_mori]
Full name
Locomotion-related protein Hikaru genki
Location in the cell
Extracellular Reliability : 2.503
Sequence
CDS
ATGCAGCTCTTGTTGCTAATATTTATTCCCTTCGTGGTCAGTGAGGACACTGGTATTGATGATTTTTCAGAAGAAGATTTTAAGTGCTCCCTACCTGATGTAACCAGTCCTAATCAAAAAGTTGATATAAATAGGATTCGTGCGGACTTCTCTAAAATATCTGAGGTTAAATTTCCTGGTTTGATTGGTCCATTAAATGAAAGATTAATCTGCAAAATTAAGTGCATCGATGGGAACTGGGTAGGACCGCTTTGCGCTAGTACTCCGGATGGTAGATTCCAACCAATATTAAGGCAGTGCTTATACAAACACGAACATCCACTATTAGCAATTTCATTTAGAAATTCTTCAATTGAAAAAGAAACTAGTTTTCCACATGGCTCTTCAATAGTGGCAAGGTGTATACATTTCGGTTTTTATAAGCTGAAAGGAGATAACACAATGCACTGTGAGAATGGGGAGTGGGTGCCAAAGTTTCCAGAATGCGTTCCGACCAGTGTCATCACTAATTTTACAGGCGACGCTCCACCCACTATACAGTACGCAATTATAAGTGGTTCAGCAGTTGTAGAACCCACAGGAGAATTGTTCGTTTTCCCTGACAGTACTCTACGTCTAGACTGTGTTACTCCAAGAGCTTTAGGCAACCCGGAATGGAGTTGGACGCAAGCCCTTGGAAATTACGAAACCGGCTGGTCATCGGACGAAGGCGAGAAGTATACCCATTACAGACTGACGCTGTCAAAGATGTCCGCGCGTCATAGTGACACGTACACTTGTGCCACTCCAACGGGACACACCAATACAGTGATCGTTAAAGTTGTTAATGTAGTTTGTCCAACAATAAACGTGTCTTCGCCTCGGCTACGTCAGTTTACACAAGGAACTAGACTTGGTCATTCGGCCCACTTTAGTTGTCAATCTGGTTTTCATCTTAATGGCTCTGCAATTTTGACTTGTATGGGAAACGGTCATTGGTCAACCAGCCTTCCGACATGCCTGGAAACTTTCTGTCCCATGTTGAAGTCTTTAGGTCCTCACTTAAGTTTCGTTGAATATAATTCGTCATTTGGGGGTCGTGCCGTATTTCAGTGTGCCTGGGGCTACAGGCTTGTGGGTTCACCAGGTTTGGAATGTGAACATGACGGTCAGTGGTCTGGAGAGGTGCCAATTTGTTCACCAATCTATTGCCCTGATCCATTAGTCCCAGAAAAGGGACGTCTGCTAATAGAGCCATCTTCAAAGCACGGCAAATATACTGTTGGGGACCTTATGATATATGATTGCGAAGACGGTTACGACATTGTCGGAGAGTCTTCTATAGTCTGTACTGAAAACGGATATTGGTCACATCCCCCACCTTTCTGCCTACTCCCTTCGGTGATCAAAAAGGCTGATACTATATTTATTGAGAATATAACATTAGTTCATGTCGAAGAATAA
Protein
MQLLLLIFIPFVVSEDTGIDDFSEEDFKCSLPDVTSPNQKVDINRIRADFSKISEVKFPGLIGPLNERLICKIKCIDGNWVGPLCASTPDGRFQPILRQCLYKHEHPLLAISFRNSSIEKETSFPHGSSIVARCIHFGFYKLKGDNTMHCENGEWVPKFPECVPTSVITNFTGDAPPTIQYAIISGSAVVEPTGELFVFPDSTLRLDCVTPRALGNPEWSWTQALGNYETGWSSDEGEKYTHYRLTLSKMSARHSDTYTCATPTGHTNTVIVKVVNVVCPTINVSSPRLRQFTQGTRLGHSAHFSCQSGFHLNGSAILTCMGNGHWSTSLPTCLETFCPMLKSLGPHLSFVEYNSSFGGRAVFQCAWGYRLVGSPGLECEHDGQWSGEVPICSPIYCPDPLVPEKGRLLIEPSSKHGKYTVGDLMIYDCEDGYDIVGESSIVCTENGYWSHPPPFCLLPSVIKKADTIFIENITLVHVEE
Summary
Description
Plays a role in the formation of functional neural circuits from the early stages of synapse formation. Has a role in the development of CNS functions involved in locomotor activity.
Keywords
Alternative splicing
Complete proteome
Developmental protein
Differentiation
Disulfide bond
Glycoprotein
Immunoglobulin domain
Neurogenesis
Reference proteome
Repeat
Secreted
Signal
Sushi
Feature
chain Locomotion-related protein Hikaru genki
splice variant In isoform 2 and isoform 4.
splice variant In isoform 2 and isoform 4.
Uniprot
Q8T107
A0A2H1VCU8
A0A2A4JSR1
A0A194QUI8
A0A194PYG6
A0A212EMP9
+ More
A0A1Y1LXX9 D6WED9 A0A182UF95 A0A182YP52 A0A067RD90 A0A182V460 A0A084VU15 A0A182L022 N6U4T2 A0A182QFQ6 A0A1S4F8F7 U4UAZ7 A0A182X1A0 A0A182VSH5 A0A097C0C8 A0A182NT73 A0A1B6DY07 A0A182IBS7 A0A182GYI5 A0A182J4C0 Q17C51 A0A1Q3G561 A0A1Q3G566 T1PM56 A0A1I8M8I0 A0A182FNB2 A0A1B0CI55 A0A1I8P321 A0A336K207 A0A1I8P364 A0A182P4N1 A0A0P8Y3Y9 A0A1Q3G549 A0A1Q3G551 A0A0P8XP02 Q09101-3 B4HSP9 A0A0Q9XJK2 T1PKT7 A0A1I8P323 A0A1I8M8H7 A0A0J9R8T5 A0A3B0JCJ9 A0A0Q5WBT7 Q09101-4 A0A0R1DMH4 A0A1W4UXL7 E0R955 A0A1A9ZVK6 A0A0M4EKS7 A0A0Q9WT91 N6W437 A0A1I8P316 A0A1I8P341 A0A1J1JBT1 A0A034VZY3 A0A3B0JK00 B3MFL4 Q09101-2 B4KPP0 A0A0L0CFV5 A0A0J9R8T6 Q09101 B3N7I7 B4P3Z6 A0A1W4UK55 A0A1B0G5E7 B4J7Q6 B4N5Z2 Q28ZI0 B4LJ78 A0A034VWD4 A0A0A1XH07 B4QH67 A0NF06 A0A1S4GZB5 A0A1S4GZ96 A0A146L2J9 A0A0M9A6Z0 J9M6V7 A0A088A3G8 A0A2H8U1F6 V9IKP1 K7IW33 E0W1Q1 A0A232FJL9 A0A1B0AZJ9 A0A1A9YPV5 A0A1A9VAB3 A0A0L7QS42 A0A154NYB2 A0A026WV69 A0A2S2QAW8
A0A1Y1LXX9 D6WED9 A0A182UF95 A0A182YP52 A0A067RD90 A0A182V460 A0A084VU15 A0A182L022 N6U4T2 A0A182QFQ6 A0A1S4F8F7 U4UAZ7 A0A182X1A0 A0A182VSH5 A0A097C0C8 A0A182NT73 A0A1B6DY07 A0A182IBS7 A0A182GYI5 A0A182J4C0 Q17C51 A0A1Q3G561 A0A1Q3G566 T1PM56 A0A1I8M8I0 A0A182FNB2 A0A1B0CI55 A0A1I8P321 A0A336K207 A0A1I8P364 A0A182P4N1 A0A0P8Y3Y9 A0A1Q3G549 A0A1Q3G551 A0A0P8XP02 Q09101-3 B4HSP9 A0A0Q9XJK2 T1PKT7 A0A1I8P323 A0A1I8M8H7 A0A0J9R8T5 A0A3B0JCJ9 A0A0Q5WBT7 Q09101-4 A0A0R1DMH4 A0A1W4UXL7 E0R955 A0A1A9ZVK6 A0A0M4EKS7 A0A0Q9WT91 N6W437 A0A1I8P316 A0A1I8P341 A0A1J1JBT1 A0A034VZY3 A0A3B0JK00 B3MFL4 Q09101-2 B4KPP0 A0A0L0CFV5 A0A0J9R8T6 Q09101 B3N7I7 B4P3Z6 A0A1W4UK55 A0A1B0G5E7 B4J7Q6 B4N5Z2 Q28ZI0 B4LJ78 A0A034VWD4 A0A0A1XH07 B4QH67 A0NF06 A0A1S4GZB5 A0A1S4GZ96 A0A146L2J9 A0A0M9A6Z0 J9M6V7 A0A088A3G8 A0A2H8U1F6 V9IKP1 K7IW33 E0W1Q1 A0A232FJL9 A0A1B0AZJ9 A0A1A9YPV5 A0A1A9VAB3 A0A0L7QS42 A0A154NYB2 A0A026WV69 A0A2S2QAW8
Pubmed
12715162
26354079
22118469
28004739
18362917
19820115
+ More
25244985 24845553 24438588 20966253 23537049 17510324 26483478 25315136 17994087 8461133 10731132 12537572 12537569 8625810 9914413 17893096 22936249 17550304 15632085 25348373 26108605 25830018 12364791 26823975 20075255 20566863 28648823 24508170
25244985 24845553 24438588 20966253 23537049 17510324 26483478 25315136 17994087 8461133 10731132 12537572 12537569 8625810 9914413 17893096 22936249 17550304 15632085 25348373 26108605 25830018 12364791 26823975 20075255 20566863 28648823 24508170
EMBL
AB079861
AB090307
BAB85192.1
ODYU01001673
SOQ38222.1
NWSH01000699
+ More
PCG74728.1 KQ461108 KPJ09193.1 KQ459594 KPI96195.1 AGBW02013811 OWR42747.1 GEZM01045068 JAV77941.1 KQ971318 EFA01313.2 KK852542 KDR21722.1 ATLV01016628 KE525098 KFB41459.1 APGK01049590 APGK01049591 KB741156 ENN73567.1 AXCN02000728 KB632169 ERL89483.1 KM363233 AIS74715.1 GEDC01006744 JAS30554.1 APCN01000734 APCN01000735 JXUM01097762 JXUM01097763 JXUM01097764 JXUM01097765 KQ564368 KXJ72377.1 CH477313 EAT43851.1 GFDL01000111 JAV34934.1 GFDL01000112 JAV34933.1 KA649160 AFP63789.1 AJWK01013041 AJWK01013042 AJWK01013043 AJWK01013044 AJWK01013045 AJWK01013046 AJWK01013047 UFQS01000031 UFQT01000031 SSW97842.1 SSX18228.1 CH902619 KPU76370.1 GFDL01000120 JAV34925.1 GFDL01000121 JAV34924.1 KPU76371.1 D13884 D13885 D13886 D13887 AE013599 AY069188 BT001427 CH480816 EDW47076.1 CH933808 KRG04471.1 KA649329 AFP63958.1 CM002911 KMY92483.1 OUUW01000001 SPP72940.1 CH954177 KQS70796.1 CM000157 KRJ98468.1 BT125623 ADM33451.1 CP012524 ALC42042.1 CH964154 KRF99156.1 CM000071 ENO01632.1 CVRI01000075 CRL08537.1 GAKP01011310 JAC47642.1 SPP72941.1 EDV36699.1 EDW09150.1 JRES01000440 KNC31126.1 KMY92482.1 EDV59392.1 EDW89479.1 CCAG010007282 CH916367 EDW02204.1 EDW79781.1 EAL25633.2 CH940648 EDW61514.2 GAKP01011311 JAC47641.1 GBXI01004055 JAD10237.1 CM000362 EDX06321.1 AAAB01008964 EAU76294.2 GDHC01016011 JAQ02618.1 KQ435724 KOX78515.1 ABLF02038631 ABLF02038632 ABLF02038638 ABLF02038639 ABLF02038645 ABLF02038646 ABLF02042181 ABLF02044375 GFXV01008086 MBW19891.1 JR049720 AEY61051.1 DS235873 EEB19633.1 NNAY01000104 OXU30931.1 JXJN01006339 JXJN01006340 JXJN01006341 KQ414758 KOC61448.1 KQ434783 KZC04666.1 KK107111 EZA58999.1 GGMS01005680 MBY74883.1
PCG74728.1 KQ461108 KPJ09193.1 KQ459594 KPI96195.1 AGBW02013811 OWR42747.1 GEZM01045068 JAV77941.1 KQ971318 EFA01313.2 KK852542 KDR21722.1 ATLV01016628 KE525098 KFB41459.1 APGK01049590 APGK01049591 KB741156 ENN73567.1 AXCN02000728 KB632169 ERL89483.1 KM363233 AIS74715.1 GEDC01006744 JAS30554.1 APCN01000734 APCN01000735 JXUM01097762 JXUM01097763 JXUM01097764 JXUM01097765 KQ564368 KXJ72377.1 CH477313 EAT43851.1 GFDL01000111 JAV34934.1 GFDL01000112 JAV34933.1 KA649160 AFP63789.1 AJWK01013041 AJWK01013042 AJWK01013043 AJWK01013044 AJWK01013045 AJWK01013046 AJWK01013047 UFQS01000031 UFQT01000031 SSW97842.1 SSX18228.1 CH902619 KPU76370.1 GFDL01000120 JAV34925.1 GFDL01000121 JAV34924.1 KPU76371.1 D13884 D13885 D13886 D13887 AE013599 AY069188 BT001427 CH480816 EDW47076.1 CH933808 KRG04471.1 KA649329 AFP63958.1 CM002911 KMY92483.1 OUUW01000001 SPP72940.1 CH954177 KQS70796.1 CM000157 KRJ98468.1 BT125623 ADM33451.1 CP012524 ALC42042.1 CH964154 KRF99156.1 CM000071 ENO01632.1 CVRI01000075 CRL08537.1 GAKP01011310 JAC47642.1 SPP72941.1 EDV36699.1 EDW09150.1 JRES01000440 KNC31126.1 KMY92482.1 EDV59392.1 EDW89479.1 CCAG010007282 CH916367 EDW02204.1 EDW79781.1 EAL25633.2 CH940648 EDW61514.2 GAKP01011311 JAC47641.1 GBXI01004055 JAD10237.1 CM000362 EDX06321.1 AAAB01008964 EAU76294.2 GDHC01016011 JAQ02618.1 KQ435724 KOX78515.1 ABLF02038631 ABLF02038632 ABLF02038638 ABLF02038639 ABLF02038645 ABLF02038646 ABLF02042181 ABLF02044375 GFXV01008086 MBW19891.1 JR049720 AEY61051.1 DS235873 EEB19633.1 NNAY01000104 OXU30931.1 JXJN01006339 JXJN01006340 JXJN01006341 KQ414758 KOC61448.1 KQ434783 KZC04666.1 KK107111 EZA58999.1 GGMS01005680 MBY74883.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000007266
UP000075902
+ More
UP000076408 UP000027135 UP000075903 UP000030765 UP000075882 UP000019118 UP000075886 UP000030742 UP000076407 UP000075920 UP000075884 UP000075840 UP000069940 UP000249989 UP000075880 UP000008820 UP000095301 UP000069272 UP000092461 UP000095300 UP000075885 UP000007801 UP000000803 UP000001292 UP000009192 UP000268350 UP000008711 UP000002282 UP000192221 UP000092445 UP000092553 UP000007798 UP000001819 UP000183832 UP000037069 UP000092444 UP000001070 UP000008792 UP000000304 UP000007062 UP000053105 UP000007819 UP000005203 UP000002358 UP000009046 UP000215335 UP000092460 UP000092443 UP000078200 UP000053825 UP000076502 UP000053097
UP000076408 UP000027135 UP000075903 UP000030765 UP000075882 UP000019118 UP000075886 UP000030742 UP000076407 UP000075920 UP000075884 UP000075840 UP000069940 UP000249989 UP000075880 UP000008820 UP000095301 UP000069272 UP000092461 UP000095300 UP000075885 UP000007801 UP000000803 UP000001292 UP000009192 UP000268350 UP000008711 UP000002282 UP000192221 UP000092445 UP000092553 UP000007798 UP000001819 UP000183832 UP000037069 UP000092444 UP000001070 UP000008792 UP000000304 UP000007062 UP000053105 UP000007819 UP000005203 UP000002358 UP000009046 UP000215335 UP000092460 UP000092443 UP000078200 UP000053825 UP000076502 UP000053097
Interpro
Gene 3D
CDD
ProteinModelPortal
Q8T107
A0A2H1VCU8
A0A2A4JSR1
A0A194QUI8
A0A194PYG6
A0A212EMP9
+ More
A0A1Y1LXX9 D6WED9 A0A182UF95 A0A182YP52 A0A067RD90 A0A182V460 A0A084VU15 A0A182L022 N6U4T2 A0A182QFQ6 A0A1S4F8F7 U4UAZ7 A0A182X1A0 A0A182VSH5 A0A097C0C8 A0A182NT73 A0A1B6DY07 A0A182IBS7 A0A182GYI5 A0A182J4C0 Q17C51 A0A1Q3G561 A0A1Q3G566 T1PM56 A0A1I8M8I0 A0A182FNB2 A0A1B0CI55 A0A1I8P321 A0A336K207 A0A1I8P364 A0A182P4N1 A0A0P8Y3Y9 A0A1Q3G549 A0A1Q3G551 A0A0P8XP02 Q09101-3 B4HSP9 A0A0Q9XJK2 T1PKT7 A0A1I8P323 A0A1I8M8H7 A0A0J9R8T5 A0A3B0JCJ9 A0A0Q5WBT7 Q09101-4 A0A0R1DMH4 A0A1W4UXL7 E0R955 A0A1A9ZVK6 A0A0M4EKS7 A0A0Q9WT91 N6W437 A0A1I8P316 A0A1I8P341 A0A1J1JBT1 A0A034VZY3 A0A3B0JK00 B3MFL4 Q09101-2 B4KPP0 A0A0L0CFV5 A0A0J9R8T6 Q09101 B3N7I7 B4P3Z6 A0A1W4UK55 A0A1B0G5E7 B4J7Q6 B4N5Z2 Q28ZI0 B4LJ78 A0A034VWD4 A0A0A1XH07 B4QH67 A0NF06 A0A1S4GZB5 A0A1S4GZ96 A0A146L2J9 A0A0M9A6Z0 J9M6V7 A0A088A3G8 A0A2H8U1F6 V9IKP1 K7IW33 E0W1Q1 A0A232FJL9 A0A1B0AZJ9 A0A1A9YPV5 A0A1A9VAB3 A0A0L7QS42 A0A154NYB2 A0A026WV69 A0A2S2QAW8
A0A1Y1LXX9 D6WED9 A0A182UF95 A0A182YP52 A0A067RD90 A0A182V460 A0A084VU15 A0A182L022 N6U4T2 A0A182QFQ6 A0A1S4F8F7 U4UAZ7 A0A182X1A0 A0A182VSH5 A0A097C0C8 A0A182NT73 A0A1B6DY07 A0A182IBS7 A0A182GYI5 A0A182J4C0 Q17C51 A0A1Q3G561 A0A1Q3G566 T1PM56 A0A1I8M8I0 A0A182FNB2 A0A1B0CI55 A0A1I8P321 A0A336K207 A0A1I8P364 A0A182P4N1 A0A0P8Y3Y9 A0A1Q3G549 A0A1Q3G551 A0A0P8XP02 Q09101-3 B4HSP9 A0A0Q9XJK2 T1PKT7 A0A1I8P323 A0A1I8M8H7 A0A0J9R8T5 A0A3B0JCJ9 A0A0Q5WBT7 Q09101-4 A0A0R1DMH4 A0A1W4UXL7 E0R955 A0A1A9ZVK6 A0A0M4EKS7 A0A0Q9WT91 N6W437 A0A1I8P316 A0A1I8P341 A0A1J1JBT1 A0A034VZY3 A0A3B0JK00 B3MFL4 Q09101-2 B4KPP0 A0A0L0CFV5 A0A0J9R8T6 Q09101 B3N7I7 B4P3Z6 A0A1W4UK55 A0A1B0G5E7 B4J7Q6 B4N5Z2 Q28ZI0 B4LJ78 A0A034VWD4 A0A0A1XH07 B4QH67 A0NF06 A0A1S4GZB5 A0A1S4GZ96 A0A146L2J9 A0A0M9A6Z0 J9M6V7 A0A088A3G8 A0A2H8U1F6 V9IKP1 K7IW33 E0W1Q1 A0A232FJL9 A0A1B0AZJ9 A0A1A9YPV5 A0A1A9VAB3 A0A0L7QS42 A0A154NYB2 A0A026WV69 A0A2S2QAW8
PDB
2Q7Z
E-value=3.91525e-16,
Score=208
Ontologies
GO
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 14,
Likelihood: 0.966571
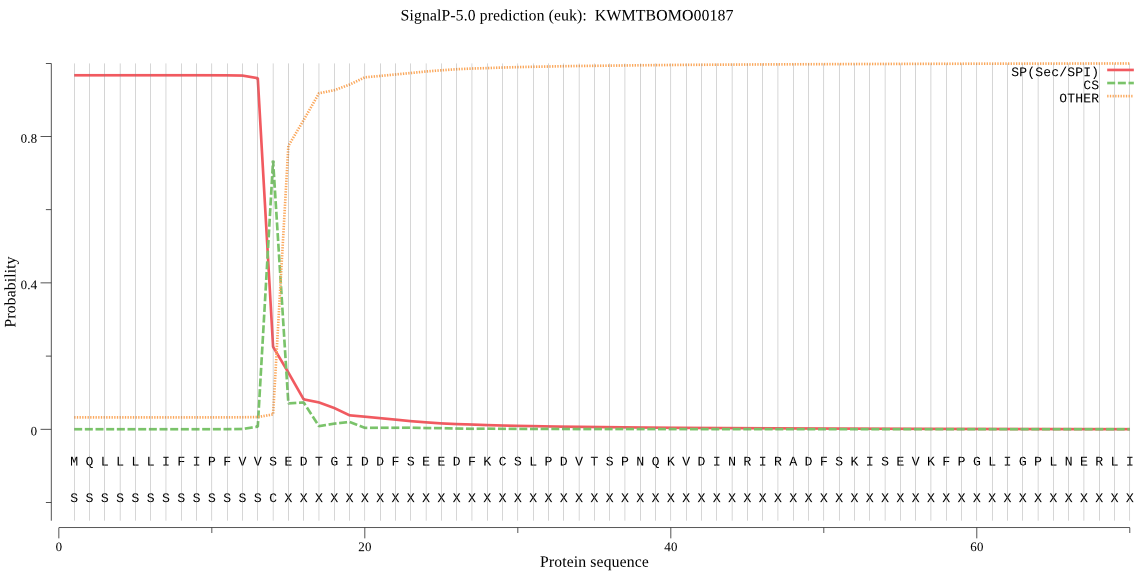
Length:
480
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00176
Exp number, first 60 AAs:
0.00068
Total prob of N-in:
0.00015
outside
1 - 480
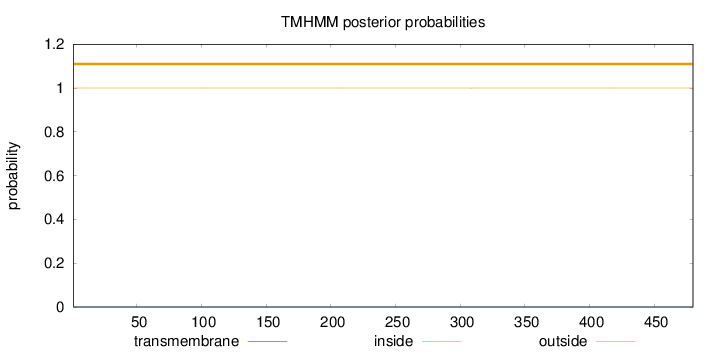
Population Genetic Test Statistics
Pi
1.082403
Theta
3.764456
Tajima's D
-2.24447
CLR
0
CSRT
0.00294985250737463
Interpretation
Uncertain
