Gene
KWMTBOMO00182
Pre Gene Modal
BGIBMGA000597
Annotation
PREDICTED:_protein_NDRG3-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.121 PlasmaMembrane Reliability : 1.092
Sequence
CDS
ATGCCATCGAACAATGCCGAAGAAACTGCTCTGCTTGGTATGGACGACATCGAACTTCGCAACATCCAACTGCAGTTCCCGAGTGCTCGCCGGTTTAGCGGAGAAGCCGCTTGTACCGAGGTGCGGGTGCGCACCGACCGCGGCGACATCCTCGTCGCTGTGCAAGGAGACCGTAACAAGCCCGCCATCATAACCTATCATGACCTCGGCCTCAACTACGCGTCATTTCAACCGTTCTTCGGATACGTCGATATGAGGGCTCTGCTCGAAAACTTTTGCGTGTACCATGTCAACGCTCCAGGCCAAGAGGAAGGCGCCCCGAGCTTGCCGGAAGACTATATGTATCCATCGATGGACGAGTTGGCGAATCAACTGAACTACGTCATGGGCCACTTCGGCATCAAGACCTTTATAGGATTCGGCGTGGGCGCTGGAGCGAACGTGCTTGCTAGATTCGCTCTTATCCACCCTAAGAAGGTAGATGCGCTGACATTGATCAACTGCACATCGAACCAGGCCGGATGGATCGAGTGGGCTTACCAGAAGATGAACACTCGGTCCCTTCGTTCCCGTGGCATGACCCAGGGCGTGCTGGACTACCTTTTGTGGCATCATTTCGGACGTTTCCCCGAAGACCGCAACCACGATCTGACCCAGATGTACAAGAACTATTTCACACGTAACGTGAACCCCAGCAACCTGTCGATGTTCATTGAGGCGTACGTGCGGCGCTCCGACCTGGGTATATGCCGTAACGCCGACACCATCAAGGTGCCGGTGTTGAACATCACGGGTGCTCTGTCACCTCACGTAGACGACACGGTGACCTTCAACGGACGTCTCAACCCGAACAATTCGACTTGGATGAAGGTGATTACTGATTTTTGTTTATTGATTGCTTAG
Protein
MPSNNAEETALLGMDDIELRNIQLQFPSARRFSGEAACTEVRVRTDRGDILVAVQGDRNKPAIITYHDLGLNYASFQPFFGYVDMRALLENFCVYHVNAPGQEEGAPSLPEDYMYPSMDELANQLNYVMGHFGIKTFIGFGVGAGANVLARFALIHPKKVDALTLINCTSNQAGWIEWAYQKMNTRSLRSRGMTQGVLDYLLWHHFGRFPEDRNHDLTQMYKNYFTRNVNPSNLSMFIEAYVRRSDLGICRNADTIKVPVLNITGALSPHVDDTVTFNGRLNPNNSTWMKVITDFCLLIA
Summary
Uniprot
A0A151WUB5
A0A195D3F7
D6WIE5
A0A151JAK7
A0A3L8E0F2
A0A195FJN0
+ More
A0A212EMP0 E2CA28 A0A154NXW5 A0A1Y1NF36 E9IGE7 A0A0C9R5X4 A0A0C9RD71 V9I8H3 V9I7Z6 A0A1Y1NDF7 H9J604 A0A0C9QIM9 V9I801 A0A0L7QUG1 A0A1B6C6P6 A0A1B6DTW4 A0A1B6CUF5 A0A195BB09 A0A067RB19 A0A026WXL0 E2AD05 V9I6T0 V9I6U9 A0A1B6HET2 V9I8W4 A0A088AS25 A0A1B6F6K5 A0A1B6JTG2 A0A1B6JFE1 A0A2A4K621 N6UI40 A0A1B6J8Q5 A0A1B6H2T0 A0A310S906 A0A212FNA1 A0A1Y1NCE7 N6UR40 A0A1Y1N9T6 N6UCG0 A0A1B6KI75 U4TSW9 A0A2J7RJQ1 A0A1B6ECJ9 A0A1B6CF76 A0A1B6DW05 A0A1B6CKS7 A0A2J7RJP6 A0A1B6DUP9 A0A1B6LBZ6 A0A1B6LPC0 A0A1B6GBT7 K7J7N9 A0A2H1V6E7 A0A1A9X3V4 D3TPL7 B3MC62 A0A0L7L547 A0A1B6H4L6 A0A0P9ADB4 A0A0L0CG12 A0A0Q9X0U4 A0A0Q9WW06 A0A0Q9WNV9 A0A0A9W699 A0A1B0BGG3 A0A1W4WSX3 A0A1L8E914 A0A0R1DRW1 A0A0P8ZMU1 A0A0Q9X8C1 A0A194RL81 A0A0P8XN35 A0A1I8P965 A0A0J9U7R4 Q8IRK7 A0A0Q5VLG5 A0A0R1DRB8 A0A0Q9XJU2 A0A1I8P9E8 A0A0Q9XHA7 A0A0Q5VZ33 A0A1I8P974 A0A0A9Y7U1 A0A1L8EBL8 A0A0J9RHQ3 Q9I7V7 B4P8Y8 A0A1L8E9V7 A0A0Q5VVL7 A0A0A9Y2A5 A0A0J9RJE4 A0A0P9BX06 A0A0J9RI55 A0A1L8ECJ9
A0A212EMP0 E2CA28 A0A154NXW5 A0A1Y1NF36 E9IGE7 A0A0C9R5X4 A0A0C9RD71 V9I8H3 V9I7Z6 A0A1Y1NDF7 H9J604 A0A0C9QIM9 V9I801 A0A0L7QUG1 A0A1B6C6P6 A0A1B6DTW4 A0A1B6CUF5 A0A195BB09 A0A067RB19 A0A026WXL0 E2AD05 V9I6T0 V9I6U9 A0A1B6HET2 V9I8W4 A0A088AS25 A0A1B6F6K5 A0A1B6JTG2 A0A1B6JFE1 A0A2A4K621 N6UI40 A0A1B6J8Q5 A0A1B6H2T0 A0A310S906 A0A212FNA1 A0A1Y1NCE7 N6UR40 A0A1Y1N9T6 N6UCG0 A0A1B6KI75 U4TSW9 A0A2J7RJQ1 A0A1B6ECJ9 A0A1B6CF76 A0A1B6DW05 A0A1B6CKS7 A0A2J7RJP6 A0A1B6DUP9 A0A1B6LBZ6 A0A1B6LPC0 A0A1B6GBT7 K7J7N9 A0A2H1V6E7 A0A1A9X3V4 D3TPL7 B3MC62 A0A0L7L547 A0A1B6H4L6 A0A0P9ADB4 A0A0L0CG12 A0A0Q9X0U4 A0A0Q9WW06 A0A0Q9WNV9 A0A0A9W699 A0A1B0BGG3 A0A1W4WSX3 A0A1L8E914 A0A0R1DRW1 A0A0P8ZMU1 A0A0Q9X8C1 A0A194RL81 A0A0P8XN35 A0A1I8P965 A0A0J9U7R4 Q8IRK7 A0A0Q5VLG5 A0A0R1DRB8 A0A0Q9XJU2 A0A1I8P9E8 A0A0Q9XHA7 A0A0Q5VZ33 A0A1I8P974 A0A0A9Y7U1 A0A1L8EBL8 A0A0J9RHQ3 Q9I7V7 B4P8Y8 A0A1L8E9V7 A0A0Q5VVL7 A0A0A9Y2A5 A0A0J9RJE4 A0A0P9BX06 A0A0J9RI55 A0A1L8ECJ9
Pubmed
EMBL
KQ982753
KYQ51225.1
KQ976885
KYN07391.1
KQ971334
EFA01066.1
+ More
KQ979302 KYN21983.1 QOIP01000002 RLU25659.1 KQ981523 KYN40209.1 AGBW02013811 OWR42739.1 GL453920 EFN75183.1 KQ434772 KZC03934.1 GEZM01009844 JAV94267.1 GL762997 EFZ20356.1 GBYB01003360 JAG73127.1 GBYB01004866 GBYB01004876 JAG74633.1 JAG74643.1 JR035902 AEY56771.1 JR035898 JR035899 AEY56767.1 AEY56768.1 GEZM01009845 JAV94266.1 BABH01029906 GBYB01003364 JAG73131.1 JR035903 AEY56772.1 KQ414736 KOC62101.1 GEDC01028224 JAS09074.1 GEDC01008213 JAS29085.1 GEDC01020176 JAS17122.1 KQ976530 KYM81718.1 KK852768 KDR16945.1 KK107069 EZA60767.1 GL438632 EFN68689.1 JR035904 AEY56773.1 JR035900 JR035901 AEY56769.1 AEY56770.1 GECU01034561 JAS73145.1 JR035905 AEY56774.1 GECZ01024276 JAS45493.1 GECU01005188 JAT02519.1 GECU01009791 GECU01004409 JAS97915.1 JAT03298.1 NWSH01000091 PCG79677.1 APGK01034449 KB740914 ENN78322.1 GECU01012142 JAS95564.1 GECZ01000839 JAS68930.1 KQ763659 OAD54893.1 AGBW02006247 OWR55224.1 GEZM01009846 JAV94265.1 APGK01005064 KB736063 ENN83261.1 GEZM01009843 JAV94268.1 APGK01034447 ENN78321.1 GEBQ01028862 JAT11115.1 KB631607 ERL84619.1 NEVH01002989 PNF41064.1 GEDC01001704 JAS35594.1 GEDC01025182 GEDC01025069 JAS12116.1 JAS12229.1 GEDC01007471 JAS29827.1 GEDC01023204 JAS14094.1 PNF41065.1 GEDC01011597 GEDC01007898 JAS25701.1 JAS29400.1 GEBQ01018768 JAT21209.1 GEBQ01014520 JAT25457.1 GECZ01009883 JAS59886.1 AAZX01002864 ODYU01000905 SOQ36369.1 EZ423369 ADD19645.1 CH902619 EDV36162.1 JTDY01002937 KOB70456.1 GECZ01000153 JAS69616.1 KPU76053.1 JRES01000440 KNC31157.1 CH963846 KRF97554.1 KRF97556.1 KRF97557.1 GBHO01040673 GBRD01002285 JAG02931.1 JAG63536.1 JXJN01013840 JXJN01013841 GFDG01003723 JAV15076.1 CM000158 KRJ99726.1 KPU76049.1 CH933808 KRG04623.1 KQ460045 KPJ18179.1 KPU76046.1 CM002911 KMY95555.1 AE013599 AAM70904.3 CH954179 KQS62295.1 KRJ99724.1 KRG04625.1 KRG04628.1 KQS62298.1 GBHO01016441 GDHC01005623 JAG27163.1 JAQ13006.1 GFDG01002793 JAV16006.1 KMY95568.1 AAM70907.2 EDW91242.1 GFDG01003413 JAV15386.1 KQS62299.1 GBHO01016437 GBRD01015544 JAG27167.1 JAG50282.1 KMY95564.1 KPU76048.1 KPU76050.1 KMY95566.1 GFDG01002331 JAV16468.1
KQ979302 KYN21983.1 QOIP01000002 RLU25659.1 KQ981523 KYN40209.1 AGBW02013811 OWR42739.1 GL453920 EFN75183.1 KQ434772 KZC03934.1 GEZM01009844 JAV94267.1 GL762997 EFZ20356.1 GBYB01003360 JAG73127.1 GBYB01004866 GBYB01004876 JAG74633.1 JAG74643.1 JR035902 AEY56771.1 JR035898 JR035899 AEY56767.1 AEY56768.1 GEZM01009845 JAV94266.1 BABH01029906 GBYB01003364 JAG73131.1 JR035903 AEY56772.1 KQ414736 KOC62101.1 GEDC01028224 JAS09074.1 GEDC01008213 JAS29085.1 GEDC01020176 JAS17122.1 KQ976530 KYM81718.1 KK852768 KDR16945.1 KK107069 EZA60767.1 GL438632 EFN68689.1 JR035904 AEY56773.1 JR035900 JR035901 AEY56769.1 AEY56770.1 GECU01034561 JAS73145.1 JR035905 AEY56774.1 GECZ01024276 JAS45493.1 GECU01005188 JAT02519.1 GECU01009791 GECU01004409 JAS97915.1 JAT03298.1 NWSH01000091 PCG79677.1 APGK01034449 KB740914 ENN78322.1 GECU01012142 JAS95564.1 GECZ01000839 JAS68930.1 KQ763659 OAD54893.1 AGBW02006247 OWR55224.1 GEZM01009846 JAV94265.1 APGK01005064 KB736063 ENN83261.1 GEZM01009843 JAV94268.1 APGK01034447 ENN78321.1 GEBQ01028862 JAT11115.1 KB631607 ERL84619.1 NEVH01002989 PNF41064.1 GEDC01001704 JAS35594.1 GEDC01025182 GEDC01025069 JAS12116.1 JAS12229.1 GEDC01007471 JAS29827.1 GEDC01023204 JAS14094.1 PNF41065.1 GEDC01011597 GEDC01007898 JAS25701.1 JAS29400.1 GEBQ01018768 JAT21209.1 GEBQ01014520 JAT25457.1 GECZ01009883 JAS59886.1 AAZX01002864 ODYU01000905 SOQ36369.1 EZ423369 ADD19645.1 CH902619 EDV36162.1 JTDY01002937 KOB70456.1 GECZ01000153 JAS69616.1 KPU76053.1 JRES01000440 KNC31157.1 CH963846 KRF97554.1 KRF97556.1 KRF97557.1 GBHO01040673 GBRD01002285 JAG02931.1 JAG63536.1 JXJN01013840 JXJN01013841 GFDG01003723 JAV15076.1 CM000158 KRJ99726.1 KPU76049.1 CH933808 KRG04623.1 KQ460045 KPJ18179.1 KPU76046.1 CM002911 KMY95555.1 AE013599 AAM70904.3 CH954179 KQS62295.1 KRJ99724.1 KRG04625.1 KRG04628.1 KQS62298.1 GBHO01016441 GDHC01005623 JAG27163.1 JAQ13006.1 GFDG01002793 JAV16006.1 KMY95568.1 AAM70907.2 EDW91242.1 GFDG01003413 JAV15386.1 KQS62299.1 GBHO01016437 GBRD01015544 JAG27167.1 JAG50282.1 KMY95564.1 KPU76048.1 KPU76050.1 KMY95566.1 GFDG01002331 JAV16468.1
Proteomes
UP000075809
UP000078542
UP000007266
UP000078492
UP000279307
UP000078541
+ More
UP000007151 UP000008237 UP000076502 UP000005204 UP000053825 UP000078540 UP000027135 UP000053097 UP000000311 UP000005203 UP000218220 UP000019118 UP000030742 UP000235965 UP000002358 UP000091820 UP000007801 UP000037510 UP000037069 UP000007798 UP000092460 UP000192223 UP000002282 UP000009192 UP000053240 UP000095300 UP000000803 UP000008711
UP000007151 UP000008237 UP000076502 UP000005204 UP000053825 UP000078540 UP000027135 UP000053097 UP000000311 UP000005203 UP000218220 UP000019118 UP000030742 UP000235965 UP000002358 UP000091820 UP000007801 UP000037510 UP000037069 UP000007798 UP000092460 UP000192223 UP000002282 UP000009192 UP000053240 UP000095300 UP000000803 UP000008711
Pfam
PF03096 Ndr
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A151WUB5
A0A195D3F7
D6WIE5
A0A151JAK7
A0A3L8E0F2
A0A195FJN0
+ More
A0A212EMP0 E2CA28 A0A154NXW5 A0A1Y1NF36 E9IGE7 A0A0C9R5X4 A0A0C9RD71 V9I8H3 V9I7Z6 A0A1Y1NDF7 H9J604 A0A0C9QIM9 V9I801 A0A0L7QUG1 A0A1B6C6P6 A0A1B6DTW4 A0A1B6CUF5 A0A195BB09 A0A067RB19 A0A026WXL0 E2AD05 V9I6T0 V9I6U9 A0A1B6HET2 V9I8W4 A0A088AS25 A0A1B6F6K5 A0A1B6JTG2 A0A1B6JFE1 A0A2A4K621 N6UI40 A0A1B6J8Q5 A0A1B6H2T0 A0A310S906 A0A212FNA1 A0A1Y1NCE7 N6UR40 A0A1Y1N9T6 N6UCG0 A0A1B6KI75 U4TSW9 A0A2J7RJQ1 A0A1B6ECJ9 A0A1B6CF76 A0A1B6DW05 A0A1B6CKS7 A0A2J7RJP6 A0A1B6DUP9 A0A1B6LBZ6 A0A1B6LPC0 A0A1B6GBT7 K7J7N9 A0A2H1V6E7 A0A1A9X3V4 D3TPL7 B3MC62 A0A0L7L547 A0A1B6H4L6 A0A0P9ADB4 A0A0L0CG12 A0A0Q9X0U4 A0A0Q9WW06 A0A0Q9WNV9 A0A0A9W699 A0A1B0BGG3 A0A1W4WSX3 A0A1L8E914 A0A0R1DRW1 A0A0P8ZMU1 A0A0Q9X8C1 A0A194RL81 A0A0P8XN35 A0A1I8P965 A0A0J9U7R4 Q8IRK7 A0A0Q5VLG5 A0A0R1DRB8 A0A0Q9XJU2 A0A1I8P9E8 A0A0Q9XHA7 A0A0Q5VZ33 A0A1I8P974 A0A0A9Y7U1 A0A1L8EBL8 A0A0J9RHQ3 Q9I7V7 B4P8Y8 A0A1L8E9V7 A0A0Q5VVL7 A0A0A9Y2A5 A0A0J9RJE4 A0A0P9BX06 A0A0J9RI55 A0A1L8ECJ9
A0A212EMP0 E2CA28 A0A154NXW5 A0A1Y1NF36 E9IGE7 A0A0C9R5X4 A0A0C9RD71 V9I8H3 V9I7Z6 A0A1Y1NDF7 H9J604 A0A0C9QIM9 V9I801 A0A0L7QUG1 A0A1B6C6P6 A0A1B6DTW4 A0A1B6CUF5 A0A195BB09 A0A067RB19 A0A026WXL0 E2AD05 V9I6T0 V9I6U9 A0A1B6HET2 V9I8W4 A0A088AS25 A0A1B6F6K5 A0A1B6JTG2 A0A1B6JFE1 A0A2A4K621 N6UI40 A0A1B6J8Q5 A0A1B6H2T0 A0A310S906 A0A212FNA1 A0A1Y1NCE7 N6UR40 A0A1Y1N9T6 N6UCG0 A0A1B6KI75 U4TSW9 A0A2J7RJQ1 A0A1B6ECJ9 A0A1B6CF76 A0A1B6DW05 A0A1B6CKS7 A0A2J7RJP6 A0A1B6DUP9 A0A1B6LBZ6 A0A1B6LPC0 A0A1B6GBT7 K7J7N9 A0A2H1V6E7 A0A1A9X3V4 D3TPL7 B3MC62 A0A0L7L547 A0A1B6H4L6 A0A0P9ADB4 A0A0L0CG12 A0A0Q9X0U4 A0A0Q9WW06 A0A0Q9WNV9 A0A0A9W699 A0A1B0BGG3 A0A1W4WSX3 A0A1L8E914 A0A0R1DRW1 A0A0P8ZMU1 A0A0Q9X8C1 A0A194RL81 A0A0P8XN35 A0A1I8P965 A0A0J9U7R4 Q8IRK7 A0A0Q5VLG5 A0A0R1DRB8 A0A0Q9XJU2 A0A1I8P9E8 A0A0Q9XHA7 A0A0Q5VZ33 A0A1I8P974 A0A0A9Y7U1 A0A1L8EBL8 A0A0J9RHQ3 Q9I7V7 B4P8Y8 A0A1L8E9V7 A0A0Q5VVL7 A0A0A9Y2A5 A0A0J9RJE4 A0A0P9BX06 A0A0J9RI55 A0A1L8ECJ9
PDB
2XMR
E-value=1.73187e-50,
Score=502
Ontologies
PANTHER
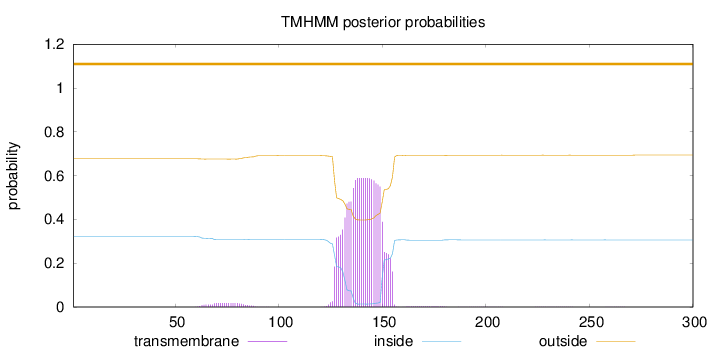
Topology
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.45496
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.32264
outside
1 - 300
Population Genetic Test Statistics
Pi
18.239919
Theta
17.854015
Tajima's D
-1.376621
CLR
0.041026
CSRT
0.0744962751862407
Interpretation
Uncertain
