Pre Gene Modal
BGIBMGA000617
Annotation
PREDICTED:_syntaxin-7_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.763
Sequence
CDS
ATGGAGGCGTCTTATCAAGGAGGTGTGTCATACGATGAGAATGGAGATAACTTTCAACGACTTTCTCAAACTATCGCCAGTAATATTAAGAAAATATCCCAAAATGTATCCTCAATGTCAAAGATGGTCAATCAGTTACAAACACCTCAAGACTCCCAAGAGCTTAGGGCTCAGTTACGCCAAATACAAAATTACACTCAGAAGCTGGCAAAGGACACTTCCTCTTTGTTAATGGAGTTGATGAGAATGCCTAAAGATAACGCTGCCAACAAACTTACCAGAGAACGACTCAGTGATGAGTATATTGCCACTCTTAATAGTTTCCAGGCAACACAACGACTCGTTGCGCAAAAGACAAAAGAAGATGTGAAAAAAGCAAAGGCTCAGTCTATAAATATTGGGGATCCTTTCGCTATGGGGAACAGTAACAAAGATTTGGTAGAGATGACCGAGACAGATGTCCGTAAGCAGGAGCAAATGAATATGCAAGCTGAACGTTCTCTCGTCGAATTGGAGGAACGCGAAAGAGATATTCGGCAACTTGAGAGTGACATACTAACCGTTAACGACATCTTCAAAGAGCTGAGTACAATGATACACGCTCAAGGCGAAGTTGTGGACTCCATCGAGTCTAGTGTTGAATACACGGCACAGAACGTCGAGAGCGCCACACAGCAACTCCGGGAGGCTGGGAATTACAAAAACAAACTGAGGAAGAAGAAAGTCTACTTGGCGATAATATTGATCATTGTGGTGTCAATAATCTTGATCGTTTTGTTCCACAACTAA
Protein
MEASYQGGVSYDENGDNFQRLSQTIASNIKKISQNVSSMSKMVNQLQTPQDSQELRAQLRQIQNYTQKLAKDTSSLLMELMRMPKDNAANKLTRERLSDEYIATLNSFQATQRLVAQKTKEDVKKAKAQSINIGDPFAMGNSNKDLVEMTETDVRKQEQMNMQAERSLVELEERERDIRQLESDILTVNDIFKELSTMIHAQGEVVDSIESSVEYTAQNVESATQQLREAGNYKNKLRKKKVYLAIILIIVVSIILIVLFHN
Summary
Similarity
Belongs to the syntaxin family.
Uniprot
H9ITN7
A0A1E1WNZ0
A0A194QV56
A0A194PS96
A0A212EML2
A0A067QSR3
+ More
A0A2J7Q3G8 A0A1W4XJB4 V5GU31 A0A1Y1N6H8 A0A139WKX6 N6UDA0 A0A0T6AYX4 A0A0M8ZTN2 A0A310SDQ6 A0A0L7QXT3 E2AAQ1 A0A154NYP5 A0A195AUA2 A0A151JY43 A0A158P0Y2 A0A151ILV4 A0A0J7KW07 A0A151WWV8 V9IGT6 A0A088A9P5 E2B860 A0A151J1H6 A0A2A3EMB5 F4WTW8 K7J6B3 A0A232F2C6 A0A1B6IBC8 A0A2S2PP25 A0A0C9RFZ4 A0A026WE94 A0A0A9YMQ2 J9JMP6 T1KRS8 A0A0P5VIF7 A0A2S2R428 C3ZCV0 A0A0P4W4F8 A0A0P6IEN0 E9GCU4 C1BQ96 G3MLG4 A0A0P4W123 A0A224XHV8 A0A0K8TNM4 A0A069DRR4 A0A087T8N0 A0A0V0G534 A0A023EM67 A0A023GHE0 A0A0V1HHE0 A0A0V1FWP4 A0A0P4WB36 Q170Z8 T1I0J8 X2B1C3 A0A023FIB3 A0A224Z9H8 A0A1E1XCG4 A0A0V0ZGB2 A0A0V0UV10 A0A0V1KPS7 A0A0V0WQ85 A0A0V0TQU1 A0A0V1P0C7 A0A0V0SHI3 A0A023FW16 A0A0V1KQW1 A0A2L2XZD4 A0A0V0TPV6 A0A0V1BBC3 A0A0V1BBE2 A0A0V0UV30 A0A1B6LHC9 L7M4I9 A0A131Z143 A0A2H8TJ99 A0A0P5E754 A0A023EPH4 A0A0V1HAN4 A0A0V1HBL9 A0A131XBR4 A0A1L8DUK6 A0A0P6BKB7 T1IH79 A0A3B4D3X5 B0X7Z0 A0A336N2K0 A0A336MWB6 A0A0V1MNI9 A0A0F8AP11 W8AT89 A0A2T7NNW7
A0A2J7Q3G8 A0A1W4XJB4 V5GU31 A0A1Y1N6H8 A0A139WKX6 N6UDA0 A0A0T6AYX4 A0A0M8ZTN2 A0A310SDQ6 A0A0L7QXT3 E2AAQ1 A0A154NYP5 A0A195AUA2 A0A151JY43 A0A158P0Y2 A0A151ILV4 A0A0J7KW07 A0A151WWV8 V9IGT6 A0A088A9P5 E2B860 A0A151J1H6 A0A2A3EMB5 F4WTW8 K7J6B3 A0A232F2C6 A0A1B6IBC8 A0A2S2PP25 A0A0C9RFZ4 A0A026WE94 A0A0A9YMQ2 J9JMP6 T1KRS8 A0A0P5VIF7 A0A2S2R428 C3ZCV0 A0A0P4W4F8 A0A0P6IEN0 E9GCU4 C1BQ96 G3MLG4 A0A0P4W123 A0A224XHV8 A0A0K8TNM4 A0A069DRR4 A0A087T8N0 A0A0V0G534 A0A023EM67 A0A023GHE0 A0A0V1HHE0 A0A0V1FWP4 A0A0P4WB36 Q170Z8 T1I0J8 X2B1C3 A0A023FIB3 A0A224Z9H8 A0A1E1XCG4 A0A0V0ZGB2 A0A0V0UV10 A0A0V1KPS7 A0A0V0WQ85 A0A0V0TQU1 A0A0V1P0C7 A0A0V0SHI3 A0A023FW16 A0A0V1KQW1 A0A2L2XZD4 A0A0V0TPV6 A0A0V1BBC3 A0A0V1BBE2 A0A0V0UV30 A0A1B6LHC9 L7M4I9 A0A131Z143 A0A2H8TJ99 A0A0P5E754 A0A023EPH4 A0A0V1HAN4 A0A0V1HBL9 A0A131XBR4 A0A1L8DUK6 A0A0P6BKB7 T1IH79 A0A3B4D3X5 B0X7Z0 A0A336N2K0 A0A336MWB6 A0A0V1MNI9 A0A0F8AP11 W8AT89 A0A2T7NNW7
Pubmed
19121390
26354079
22118469
24845553
28004739
18362917
+ More
19820115 23537049 20798317 21347285 21719571 20075255 28648823 24508170 30249741 25401762 26823975 18563158 21292972 22216098 27129103 26369729 26334808 24945155 17510324 23254933 28797301 28503490 26561354 25576852 26830274 25835551 24495485
19820115 23537049 20798317 21347285 21719571 20075255 28648823 24508170 30249741 25401762 26823975 18563158 21292972 22216098 27129103 26369729 26334808 24945155 17510324 23254933 28797301 28503490 26561354 25576852 26830274 25835551 24495485
EMBL
BABH01006733
GDQN01002352
GDQN01002338
JAT88702.1
JAT88716.1
KQ461108
+ More
KPJ09199.1 KQ459594 KPI96187.1 AGBW02013811 OWR42740.1 KK852982 KDR12892.1 NEVH01019067 PNF23117.1 GALX01003314 JAB65152.1 GEZM01011431 JAV93522.1 KQ971321 KYB28649.1 APGK01033063 KB740860 KB632287 ENN78591.1 ERL91476.1 LJIG01022472 KRT80399.1 KQ435891 KOX69403.1 KQ767283 OAD53433.1 KQ414699 KOC63428.1 GL438137 EFN69501.1 KQ434783 KZC04795.1 KQ976738 KYM75756.1 KQ981515 KYN40767.1 ADTU01005902 KQ977086 KYN05868.1 LBMM01002715 KMQ94444.1 KQ982691 KYQ52145.1 JR042943 AEY59696.1 GL446283 EFN88122.1 KQ980514 KYN15746.1 KZ288212 PBC32830.1 GL888345 EGI62325.1 NNAY01001263 OXU24567.1 GECU01023460 JAS84246.1 GGMR01018554 MBY31173.1 GBYB01005975 GBYB01005976 JAG75742.1 JAG75743.1 KK107260 QOIP01000001 EZA54248.1 RLU26240.1 GBHO01010140 GBRD01006280 GDHC01000016 JAG33464.1 JAG59541.1 JAQ18613.1 ABLF02031790 CAEY01000418 GDIP01099513 LRGB01000944 JAM04202.1 KZS14524.1 GGMS01015532 MBY84735.1 GG666610 EEN49603.1 GDRN01073064 JAI63450.1 GDIQ01005647 JAN89090.1 GL732539 EFX82617.1 BT076775 ACO11199.1 JO842715 AEO34332.1 GDKW01001204 JAI55391.1 GFTR01004350 JAW12076.1 GDAI01001611 JAI15992.1 GBGD01002533 JAC86356.1 KK113970 KFM61469.1 GECL01003332 JAP02792.1 GAPW01003200 JAC10398.1 GBBM01002036 JAC33382.1 JYDU01000082 JYDR01000072 JYDS01000373 JYDV01000038 KRX93779.1 KRY70519.1 KRZ09878.1 KRZ39157.1 JYDT01000022 KRY90361.1 GDRN01073065 JAI63449.1 CH477462 EAT40546.1 ACPB03008738 AMQN01000222 GBBK01003230 JAC21252.1 GFPF01012208 MAA23354.1 GFAC01002387 JAT96801.1 JYDQ01000191 KRY11529.1 JYDN01000153 KRX55131.1 JYDW01000324 KRZ49247.1 JYDK01000076 KRX77824.1 JYDJ01000183 KRX40999.1 JYDM01000067 KRZ89621.1 JYDL01000008 KRX26255.1 GBBL01001626 JAC25694.1 KRZ49246.1 IAAA01005100 IAAA01005101 LAA01293.1 KRX41000.1 JYDH01000069 KRY34299.1 KRY34298.1 KRX55132.1 GEBQ01027684 GEBQ01016993 GEBQ01009207 JAT12293.1 JAT22984.1 JAT30770.1 GACK01005793 JAA59241.1 GEDV01003919 JAP84638.1 GFXV01001957 MBW13762.1 GDIP01151799 JAJ71603.1 GAPW01003201 GAPW01003199 JAC10399.1 JYDP01000096 KRZ07800.1 KRZ07801.1 GEFH01004544 JAP64037.1 GFDF01003963 JAV10121.1 GDIQ01222405 GDIP01027968 JAK29320.1 JAM75747.1 JH429784 DS232468 EDS42209.1 UFQT01003469 SSX35027.1 UFQT01001373 SSX30238.1 JYDO01000064 KRZ73393.1 KQ041569 KKF25197.1 GAMC01014541 GAMC01014539 GAMC01014538 GAMC01014536 GAMC01014535 JAB92020.1 PZQS01000010 PVD22842.1
KPJ09199.1 KQ459594 KPI96187.1 AGBW02013811 OWR42740.1 KK852982 KDR12892.1 NEVH01019067 PNF23117.1 GALX01003314 JAB65152.1 GEZM01011431 JAV93522.1 KQ971321 KYB28649.1 APGK01033063 KB740860 KB632287 ENN78591.1 ERL91476.1 LJIG01022472 KRT80399.1 KQ435891 KOX69403.1 KQ767283 OAD53433.1 KQ414699 KOC63428.1 GL438137 EFN69501.1 KQ434783 KZC04795.1 KQ976738 KYM75756.1 KQ981515 KYN40767.1 ADTU01005902 KQ977086 KYN05868.1 LBMM01002715 KMQ94444.1 KQ982691 KYQ52145.1 JR042943 AEY59696.1 GL446283 EFN88122.1 KQ980514 KYN15746.1 KZ288212 PBC32830.1 GL888345 EGI62325.1 NNAY01001263 OXU24567.1 GECU01023460 JAS84246.1 GGMR01018554 MBY31173.1 GBYB01005975 GBYB01005976 JAG75742.1 JAG75743.1 KK107260 QOIP01000001 EZA54248.1 RLU26240.1 GBHO01010140 GBRD01006280 GDHC01000016 JAG33464.1 JAG59541.1 JAQ18613.1 ABLF02031790 CAEY01000418 GDIP01099513 LRGB01000944 JAM04202.1 KZS14524.1 GGMS01015532 MBY84735.1 GG666610 EEN49603.1 GDRN01073064 JAI63450.1 GDIQ01005647 JAN89090.1 GL732539 EFX82617.1 BT076775 ACO11199.1 JO842715 AEO34332.1 GDKW01001204 JAI55391.1 GFTR01004350 JAW12076.1 GDAI01001611 JAI15992.1 GBGD01002533 JAC86356.1 KK113970 KFM61469.1 GECL01003332 JAP02792.1 GAPW01003200 JAC10398.1 GBBM01002036 JAC33382.1 JYDU01000082 JYDR01000072 JYDS01000373 JYDV01000038 KRX93779.1 KRY70519.1 KRZ09878.1 KRZ39157.1 JYDT01000022 KRY90361.1 GDRN01073065 JAI63449.1 CH477462 EAT40546.1 ACPB03008738 AMQN01000222 GBBK01003230 JAC21252.1 GFPF01012208 MAA23354.1 GFAC01002387 JAT96801.1 JYDQ01000191 KRY11529.1 JYDN01000153 KRX55131.1 JYDW01000324 KRZ49247.1 JYDK01000076 KRX77824.1 JYDJ01000183 KRX40999.1 JYDM01000067 KRZ89621.1 JYDL01000008 KRX26255.1 GBBL01001626 JAC25694.1 KRZ49246.1 IAAA01005100 IAAA01005101 LAA01293.1 KRX41000.1 JYDH01000069 KRY34299.1 KRY34298.1 KRX55132.1 GEBQ01027684 GEBQ01016993 GEBQ01009207 JAT12293.1 JAT22984.1 JAT30770.1 GACK01005793 JAA59241.1 GEDV01003919 JAP84638.1 GFXV01001957 MBW13762.1 GDIP01151799 JAJ71603.1 GAPW01003201 GAPW01003199 JAC10399.1 JYDP01000096 KRZ07800.1 KRZ07801.1 GEFH01004544 JAP64037.1 GFDF01003963 JAV10121.1 GDIQ01222405 GDIP01027968 JAK29320.1 JAM75747.1 JH429784 DS232468 EDS42209.1 UFQT01003469 SSX35027.1 UFQT01001373 SSX30238.1 JYDO01000064 KRZ73393.1 KQ041569 KKF25197.1 GAMC01014541 GAMC01014539 GAMC01014538 GAMC01014536 GAMC01014535 JAB92020.1 PZQS01000010 PVD22842.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000027135
UP000235965
+ More
UP000192223 UP000007266 UP000019118 UP000030742 UP000053105 UP000053825 UP000000311 UP000076502 UP000078540 UP000078541 UP000005205 UP000078542 UP000036403 UP000075809 UP000005203 UP000008237 UP000078492 UP000242457 UP000007755 UP000002358 UP000215335 UP000053097 UP000279307 UP000007819 UP000015104 UP000076858 UP000001554 UP000000305 UP000054359 UP000054632 UP000054805 UP000054815 UP000054826 UP000054995 UP000008820 UP000015103 UP000054783 UP000054681 UP000054721 UP000054673 UP000055048 UP000054924 UP000054630 UP000054776 UP000055024 UP000261440 UP000002320 UP000054843 UP000245119
UP000192223 UP000007266 UP000019118 UP000030742 UP000053105 UP000053825 UP000000311 UP000076502 UP000078540 UP000078541 UP000005205 UP000078542 UP000036403 UP000075809 UP000005203 UP000008237 UP000078492 UP000242457 UP000007755 UP000002358 UP000215335 UP000053097 UP000279307 UP000007819 UP000015104 UP000076858 UP000001554 UP000000305 UP000054359 UP000054632 UP000054805 UP000054815 UP000054826 UP000054995 UP000008820 UP000015103 UP000054783 UP000054681 UP000054721 UP000054673 UP000055048 UP000054924 UP000054630 UP000054776 UP000055024 UP000261440 UP000002320 UP000054843 UP000245119
PRIDE
Interpro
CDD
ProteinModelPortal
H9ITN7
A0A1E1WNZ0
A0A194QV56
A0A194PS96
A0A212EML2
A0A067QSR3
+ More
A0A2J7Q3G8 A0A1W4XJB4 V5GU31 A0A1Y1N6H8 A0A139WKX6 N6UDA0 A0A0T6AYX4 A0A0M8ZTN2 A0A310SDQ6 A0A0L7QXT3 E2AAQ1 A0A154NYP5 A0A195AUA2 A0A151JY43 A0A158P0Y2 A0A151ILV4 A0A0J7KW07 A0A151WWV8 V9IGT6 A0A088A9P5 E2B860 A0A151J1H6 A0A2A3EMB5 F4WTW8 K7J6B3 A0A232F2C6 A0A1B6IBC8 A0A2S2PP25 A0A0C9RFZ4 A0A026WE94 A0A0A9YMQ2 J9JMP6 T1KRS8 A0A0P5VIF7 A0A2S2R428 C3ZCV0 A0A0P4W4F8 A0A0P6IEN0 E9GCU4 C1BQ96 G3MLG4 A0A0P4W123 A0A224XHV8 A0A0K8TNM4 A0A069DRR4 A0A087T8N0 A0A0V0G534 A0A023EM67 A0A023GHE0 A0A0V1HHE0 A0A0V1FWP4 A0A0P4WB36 Q170Z8 T1I0J8 X2B1C3 A0A023FIB3 A0A224Z9H8 A0A1E1XCG4 A0A0V0ZGB2 A0A0V0UV10 A0A0V1KPS7 A0A0V0WQ85 A0A0V0TQU1 A0A0V1P0C7 A0A0V0SHI3 A0A023FW16 A0A0V1KQW1 A0A2L2XZD4 A0A0V0TPV6 A0A0V1BBC3 A0A0V1BBE2 A0A0V0UV30 A0A1B6LHC9 L7M4I9 A0A131Z143 A0A2H8TJ99 A0A0P5E754 A0A023EPH4 A0A0V1HAN4 A0A0V1HBL9 A0A131XBR4 A0A1L8DUK6 A0A0P6BKB7 T1IH79 A0A3B4D3X5 B0X7Z0 A0A336N2K0 A0A336MWB6 A0A0V1MNI9 A0A0F8AP11 W8AT89 A0A2T7NNW7
A0A2J7Q3G8 A0A1W4XJB4 V5GU31 A0A1Y1N6H8 A0A139WKX6 N6UDA0 A0A0T6AYX4 A0A0M8ZTN2 A0A310SDQ6 A0A0L7QXT3 E2AAQ1 A0A154NYP5 A0A195AUA2 A0A151JY43 A0A158P0Y2 A0A151ILV4 A0A0J7KW07 A0A151WWV8 V9IGT6 A0A088A9P5 E2B860 A0A151J1H6 A0A2A3EMB5 F4WTW8 K7J6B3 A0A232F2C6 A0A1B6IBC8 A0A2S2PP25 A0A0C9RFZ4 A0A026WE94 A0A0A9YMQ2 J9JMP6 T1KRS8 A0A0P5VIF7 A0A2S2R428 C3ZCV0 A0A0P4W4F8 A0A0P6IEN0 E9GCU4 C1BQ96 G3MLG4 A0A0P4W123 A0A224XHV8 A0A0K8TNM4 A0A069DRR4 A0A087T8N0 A0A0V0G534 A0A023EM67 A0A023GHE0 A0A0V1HHE0 A0A0V1FWP4 A0A0P4WB36 Q170Z8 T1I0J8 X2B1C3 A0A023FIB3 A0A224Z9H8 A0A1E1XCG4 A0A0V0ZGB2 A0A0V0UV10 A0A0V1KPS7 A0A0V0WQ85 A0A0V0TQU1 A0A0V1P0C7 A0A0V0SHI3 A0A023FW16 A0A0V1KQW1 A0A2L2XZD4 A0A0V0TPV6 A0A0V1BBC3 A0A0V1BBE2 A0A0V0UV30 A0A1B6LHC9 L7M4I9 A0A131Z143 A0A2H8TJ99 A0A0P5E754 A0A023EPH4 A0A0V1HAN4 A0A0V1HBL9 A0A131XBR4 A0A1L8DUK6 A0A0P6BKB7 T1IH79 A0A3B4D3X5 B0X7Z0 A0A336N2K0 A0A336MWB6 A0A0V1MNI9 A0A0F8AP11 W8AT89 A0A2T7NNW7
PDB
2DNX
E-value=1.09602e-10,
Score=158
Ontologies
PATHWAY
GO
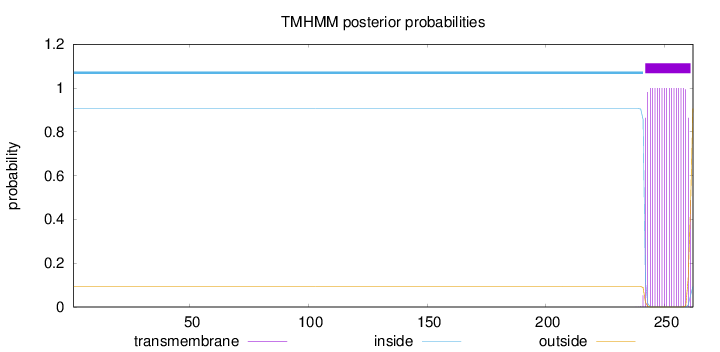
Topology
Length:
262
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.24694
Exp number, first 60 AAs:
0.00011
Total prob of N-in:
0.90659
inside
1 - 241
TMhelix
242 - 261
outside
262 - 262
Population Genetic Test Statistics
Pi
15.749812
Theta
13.179993
Tajima's D
0.086836
CLR
0.008134
CSRT
0.399730013499325
Interpretation
Uncertain
