Gene
KWMTBOMO00180
Pre Gene Modal
BGIBMGA000599
Annotation
PREDICTED:_PAB-dependent_poly(A)-specific_ribonuclease_subunit_PAN3_[Amyelois_transitella]
Full name
PAN2-PAN3 deadenylation complex subunit PAN3
Alternative Name
PAB1P-dependent poly(A)-specific ribonuclease
Poly(A)-nuclease deadenylation complex subunit 3
Poly(A)-nuclease deadenylation complex subunit 3
Location in the cell
Nuclear Reliability : 1.774
Sequence
CDS
ATGAGTCGCTCAACTTCGACGCCCACACGCTCTCTTAACCATGCTATAGGGAAGCTCACTTTGGATTCCTCACCACCAGGTGTTAAGAAGGTATTGGTGAGCGAATTTATTCCGATGAACTACTATGTTCCGCCACCGAATATGCTTCCCCCAGAATCACCTGAGGGTCCACCACCACCTCCTATACCAATGGCGAATGCTTCAACTGTCCATCAGGAGAACGTGGGAGGAACTACCTATTTCTACTCGACCAATTCAGACAGCCTCAATACCACTAGCGGTCTTAATTCATCGGCTTCCATGGAAGAACCGTGCATGGACTACTCTCTGCTGAGCACCGCAAGACCATATCCATTGGTATACCCATATTTCCACCAACCAGGTCCGTCTGAGCCATGTCTAAACGCAGTAGGGCCGCCGATGGTGTCTCCGTATGCTTCGCAGATGTACGCTGGGGCCTTACCGCAGCCAGGGCCGTCCACCAACAAACAAGGTCTAGCGGCGACTTTTTACAATCCCGAAACAATACGGTCTGAGATCTACGACAGGAATGATGATGTTTATTTGCAGCCTGATCTCAATCAGTTTCCAGATATACCGGACAGCGTGGACATGTACGCCGAGCTGGTACCCCTGGAGGGTGCCGTGACGCATTCGATGTCGACATCGTACCGCGCTACTAACCGTCAGAACGGTGATTATGTGGCTCTAAGGAGGCTCCATTCTTACACGTCGCCGGCCTCCAAGCGTCTTGAAATGTGGAAGCAGATCGACCACCCGAACATTGTGCGTCTGGAAGAATATTTCAGCACCAAGGCTTTTAACGATATATCACTGGTCCTGGTGTACCAGTATCATCCAGCCGCTGTGACTCTGATGAACAAGTATTTGTCCGGAAACGGAAACAACTCCGCCGAGGGTGGAGCTGCTTATCATGATCCGTTCTCTTCGGACCCGGATGCACCGCGTCCGTATACCCATCAGAAAAATGCCATGCTCCGTGCGGTCGCATGCGGTGCTTTGCTTCCGGAAGCTGTGTTGTGGAGTCTGCTCGTGCAACTGACGGCCGGCTTGCGCGCCATACATACAGCCGGACTCGCCTGCAGGAGCCTGGAGCCGACCAAGGTGATCATGAACGGATGTCGAGTCCGAATTGCATGGTGCGGTGCTGCTGATGCTCTACACAGCAACACCAATGATGTAGTACAGGCTCAGCAAGACGATCTGACCGCTTTGGGCCGGCTGGCACTGGCTCTAGCCTGTCGCACCATACATTGCGACAACCTCGCAGCGTCCATGGAACTCGTCGCTAGGACCTACTCTGCGGATCTGAAGAACCTAATTCTGTATCTGCTGTCATCCTCGCCTGCGCGTCGTTCCGTTATCGACCTTATGCCTATGATCGGAGCTAGGTTCTATACTCAGGTGGAGGCACTGGAGAGACGCGCTGATGCGTTTGAAGATCAGCTGTCCCAGGAGATAGATAACGGTCGTCTCCTCCGCATATTAATCAAGCTTGGCATTGTCAACGAGCGTCCAGAATTGAATCTGGATCCTGCTTGGTCGGAGACCGGAGATCGCTACATGCTGAAGCTGTTCCGTGATTATCTGTTCCATTCAGTGACGACTGACGGCCGTCCTTGGCTGGACCAAGCGCACCTAACGCACTGCCTGAACCAGCTCGACGGAGGAACTGCTGCGAAGGTGGAGCTGATGTCTCGCGACGAGCAAAGCGTCCTGGTCGTGTCCTACGCTGAGCTGAAGCACTGCCTCGACCAAGCCTTCGAGGAGCCAGAGCCCGAGCCGGAATACGGCTCCGGCTACACCAATAGCATACCCATCAGGCACGGCGACGACCTGGGCTACGTGACCAACGCGTACGGCTTCAGCTTTAGTTTCGGTTATATCAACGGGTTCAGCAGGGAGAACAACGTTCCAATAAACAATCAGTTTAATGAGCGCGACAACAACGGCGACGGCGGTAATGGCGACAACGGCGAATACTTCTTTGGCGGCCCAGGTAATGGCAACCGCTGCAGTTGCGGCCACGATTACGAATACATGGAGTAG
Protein
MSRSTSTPTRSLNHAIGKLTLDSSPPGVKKVLVSEFIPMNYYVPPPNMLPPESPEGPPPPPIPMANASTVHQENVGGTTYFYSTNSDSLNTTSGLNSSASMEEPCMDYSLLSTARPYPLVYPYFHQPGPSEPCLNAVGPPMVSPYASQMYAGALPQPGPSTNKQGLAATFYNPETIRSEIYDRNDDVYLQPDLNQFPDIPDSVDMYAELVPLEGAVTHSMSTSYRATNRQNGDYVALRRLHSYTSPASKRLEMWKQIDHPNIVRLEEYFSTKAFNDISLVLVYQYHPAAVTLMNKYLSGNGNNSAEGGAAYHDPFSSDPDAPRPYTHQKNAMLRAVACGALLPEAVLWSLLVQLTAGLRAIHTAGLACRSLEPTKVIMNGCRVRIAWCGAADALHSNTNDVVQAQQDDLTALGRLALALACRTIHCDNLAASMELVARTYSADLKNLILYLLSSSPARRSVIDLMPMIGARFYTQVEALERRADAFEDQLSQEIDNGRLLRILIKLGIVNERPELNLDPAWSETGDRYMLKLFRDYLFHSVTTDGRPWLDQAHLTHCLNQLDGGTAAKVELMSRDEQSVLVVSYAELKHCLDQAFEEPEPEPEYGSGYTNSIPIRHGDDLGYVTNAYGFSFSFGYINGFSRENNVPINNQFNERDNNGDGGNGDNGEYFFGGPGNGNRCSCGHDYEYME
Summary
Description
Regulatory subunit of the poly(A)-nuclease (PAN) deadenylation complex, one of two cytoplasmic mRNA deadenylases involved in general and miRNA-mediated mRNA turnover. PAN specifically shortens poly(A) tails of RNA and the activity is stimulated by poly(A)-binding protein (PABP). PAN deadenylation is followed by rapid degradation of the shortened mRNA tails by the CCR4-NOT complex. Deadenylated mRNAs are then degraded by two alternative mechanisms, namely exosome-mediated 3'-5' exonucleolytic degradation, or deadenlyation-dependent mRNA decaping and subsequent 5'-3' exonucleolytic degradation by XRN1. PAN3 acts as a positive regulator for PAN activity, recruiting the catalytic subunit PAN2 to mRNA via its interaction with RNA and PABP, and to miRNA targets via its interaction with GW182 family proteins.
Subunit
Homodimer. Forms a heterotrimer with a catalytic subunit PAN2 to form the poly(A)-nuclease (PAN) deadenylation complex. Interacts (via PAM-2 motif) with poly(A)-binding protein (via PABC domain), conferring substrate specificity of the enzyme complex.
Homodimer. Forms a heterotrimer with a catalytic subunit PAN2 to form the poly(A)-nuclease (PAN) deadenylation complex. Interacts (via PAM-2 motif) with poly(A)-binding protein (via PABC domain), conferring substrate specificity of the enzyme complex (PubMed:23932717). Interacts with the GW182 family protein gw (PubMed:21981923, PubMed:23172285).
Homodimer. Forms a heterotrimer with a catalytic subunit PAN2 to form the poly(A)-nuclease (PAN) deadenylation complex. Interacts (via PAM-2 motif) with poly(A)-binding protein (via PABC domain), conferring substrate specificity of the enzyme complex (PubMed:23932717). Interacts with the GW182 family protein gw (PubMed:21981923, PubMed:23172285).
Similarity
Belongs to the protein kinase superfamily. PAN3 family.
Keywords
3D-structure
ATP-binding
Coiled coil
Complete proteome
Cytoplasm
mRNA processing
Nucleotide-binding
Reference proteome
Feature
chain PAN2-PAN3 deadenylation complex subunit PAN3
Uniprot
H9ITL9
A0A2H1V790
A0A2A4K171
A0A2A4JZI1
A0A1E1WSE2
A0A194PRW4
+ More
A0A212EMS4 A0A1E1WHJ0 A0A2A4K041 A0A2A4K0H3 A0A1B6C5K5 K7J653 A0A2J7R474 A0A195BX79 A0A026W4B0 A0A195CAS4 A0A2J7R467 F4WJC7 A0A158NE99 E2A0A4 A0A310SAN1 E2B645 A0A0L7R833 A0A151WSG5 A0A195FQU3 A0A088A552 A0A151J0C7 A0A1I8PZ41 A0A1B6K0L5 E9J1J2 A0A1B6IQW3 A0A0M9A5G5 A0A1I8PZ25 A0A0L0CAC0 A0A1Y1L9N6 A0A034V5D0 A0A1B6MBR2 A0A0K8V1P7 A0A0A1XHV3 A0A154P8V5 A0A0K8V0I2 A0A2A3E6M8 T1P9Y8 A0A0R1DWT5 A0A0R1E1H7 B4PCV7 A0A1L8DXV8 A0A1L8DXP7 A0A0C9R933 A0A0J9RPD0 Q9I7T8 B4IZP5 A0A0J9RMW2 A8JNJ2 A0A0J9RNB8 A0A1I8NAT2 B4QP17 A0A0J9RMV5 A0A0Q5U287 Q7KV83 Q8IRE7 A0A0Q5UH76 A0A1I8NAU6 A0A0J9RNC2 Q95RR8 A0A0Q5U300 Q9VZV3 A0A1I8NAU1 A0A1I8NAT9 B4HTJ5 B3NF26 A0A0N8P0W3 A0A0P8ZUK3 D3DMN9 A0A0P9C3J6 B3M7E6 A0A1W4W1Q9 A0A1W4VNY6 A0A1W4VNP1 A0A1W4W118 A0A3B0JXG1 A0A3B0JBJ9 B4K3A2 Q17PM7 A0A3B0JPK7 A0A0R3P8J3 A0A0R3P6E2 A0A0M4EXP0 A0A0R3P8Z1 Q29E31 A0A0R3P6D7 B4N6I6 A0A0R3P962
A0A212EMS4 A0A1E1WHJ0 A0A2A4K041 A0A2A4K0H3 A0A1B6C5K5 K7J653 A0A2J7R474 A0A195BX79 A0A026W4B0 A0A195CAS4 A0A2J7R467 F4WJC7 A0A158NE99 E2A0A4 A0A310SAN1 E2B645 A0A0L7R833 A0A151WSG5 A0A195FQU3 A0A088A552 A0A151J0C7 A0A1I8PZ41 A0A1B6K0L5 E9J1J2 A0A1B6IQW3 A0A0M9A5G5 A0A1I8PZ25 A0A0L0CAC0 A0A1Y1L9N6 A0A034V5D0 A0A1B6MBR2 A0A0K8V1P7 A0A0A1XHV3 A0A154P8V5 A0A0K8V0I2 A0A2A3E6M8 T1P9Y8 A0A0R1DWT5 A0A0R1E1H7 B4PCV7 A0A1L8DXV8 A0A1L8DXP7 A0A0C9R933 A0A0J9RPD0 Q9I7T8 B4IZP5 A0A0J9RMW2 A8JNJ2 A0A0J9RNB8 A0A1I8NAT2 B4QP17 A0A0J9RMV5 A0A0Q5U287 Q7KV83 Q8IRE7 A0A0Q5UH76 A0A1I8NAU6 A0A0J9RNC2 Q95RR8 A0A0Q5U300 Q9VZV3 A0A1I8NAU1 A0A1I8NAT9 B4HTJ5 B3NF26 A0A0N8P0W3 A0A0P8ZUK3 D3DMN9 A0A0P9C3J6 B3M7E6 A0A1W4W1Q9 A0A1W4VNY6 A0A1W4VNP1 A0A1W4W118 A0A3B0JXG1 A0A3B0JBJ9 B4K3A2 Q17PM7 A0A3B0JPK7 A0A0R3P8J3 A0A0R3P6E2 A0A0M4EXP0 A0A0R3P8Z1 Q29E31 A0A0R3P6D7 B4N6I6 A0A0R3P962
Pubmed
19121390
26354079
22118469
20075255
24508170
30249741
+ More
21719571 21347285 20798317 21282665 26108605 28004739 25348373 25830018 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021 12537569 21981923 23172285 23932717 17510324 15632085
21719571 21347285 20798317 21282665 26108605 28004739 25348373 25830018 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 18057021 12537569 21981923 23172285 23932717 17510324 15632085
EMBL
BABH01006731
BABH01006732
ODYU01000800
SOQ36124.1
NWSH01000326
PCG77420.1
+ More
PCG77421.1 GDQN01007940 GDQN01001323 JAT83114.1 JAT89731.1 KQ459594 KPI96186.1 AGBW02013811 OWR42741.1 GDQN01004688 GDQN01000118 JAT86366.1 JAT90936.1 PCG77419.1 PCG77418.1 GEDC01028515 JAS08783.1 NEVH01007815 PNF35617.1 KQ976401 KYM92568.1 KK107455 QOIP01000010 EZA50436.1 RLU17387.1 KQ978023 KYM97912.1 PNF35616.1 GL888182 EGI65705.1 ADTU01013181 GL435569 EFN73107.1 KQ777311 OAD52141.1 GL445914 EFN88834.1 KQ414633 KOC67040.1 KQ982769 KYQ50849.1 KQ981335 KYN42667.1 KQ980636 KYN14948.1 GECU01002697 JAT05010.1 GL767671 EFZ13318.1 GECU01018384 JAS89322.1 KQ435729 KOX77625.1 JRES01000681 KNC29197.1 GEZM01066554 GEZM01066551 JAV67807.1 GAKP01020463 GAKP01020461 GAKP01020455 GAKP01020451 JAC38501.1 GEBQ01006620 JAT33357.1 GDHF01021218 GDHF01019784 GDHF01004989 JAI31096.1 JAI32530.1 JAI47325.1 GBXI01003755 JAD10537.1 KQ434846 KZC08356.1 GDHF01019900 JAI32414.1 KZ288364 PBC26876.1 KA645547 AFP60176.1 CM000159 KRK01461.1 KRK01458.1 EDW93861.1 GFDF01002803 JAV11281.1 GFDF01002854 JAV11230.1 GBYB01012884 JAG82651.1 CM002912 KMY97239.1 AE014296 BT100048 AAG22234.1 ACX61589.1 CH916366 EDV95630.1 KMY97238.1 ABW08446.1 KMY97232.1 CM000363 EDX09012.1 KMY97230.1 KMY97233.1 CH954178 KQS43231.1 BT125980 AAN11524.1 AAS64952.1 ADX35959.1 AAN11526.1 KQS43226.1 KMY97237.1 AY061183 KQS43227.1 AAF47713.3 CH480817 EDW50266.1 EDV50368.1 CH902618 KPU78244.1 KPU78243.1 BT120205 ADB91453.1 KPU78242.1 EDV39844.1 OUUW01000002 SPP77411.1 SPP77412.1 CH921324 EDV90450.1 CH477190 EAT48725.1 SPP77410.1 CH379070 KRT08719.1 KRT08722.1 CP012525 ALC43112.1 KRT08723.1 EAL30232.2 KRT08721.1 CH964161 EDW79975.2 KRT08720.1
PCG77421.1 GDQN01007940 GDQN01001323 JAT83114.1 JAT89731.1 KQ459594 KPI96186.1 AGBW02013811 OWR42741.1 GDQN01004688 GDQN01000118 JAT86366.1 JAT90936.1 PCG77419.1 PCG77418.1 GEDC01028515 JAS08783.1 NEVH01007815 PNF35617.1 KQ976401 KYM92568.1 KK107455 QOIP01000010 EZA50436.1 RLU17387.1 KQ978023 KYM97912.1 PNF35616.1 GL888182 EGI65705.1 ADTU01013181 GL435569 EFN73107.1 KQ777311 OAD52141.1 GL445914 EFN88834.1 KQ414633 KOC67040.1 KQ982769 KYQ50849.1 KQ981335 KYN42667.1 KQ980636 KYN14948.1 GECU01002697 JAT05010.1 GL767671 EFZ13318.1 GECU01018384 JAS89322.1 KQ435729 KOX77625.1 JRES01000681 KNC29197.1 GEZM01066554 GEZM01066551 JAV67807.1 GAKP01020463 GAKP01020461 GAKP01020455 GAKP01020451 JAC38501.1 GEBQ01006620 JAT33357.1 GDHF01021218 GDHF01019784 GDHF01004989 JAI31096.1 JAI32530.1 JAI47325.1 GBXI01003755 JAD10537.1 KQ434846 KZC08356.1 GDHF01019900 JAI32414.1 KZ288364 PBC26876.1 KA645547 AFP60176.1 CM000159 KRK01461.1 KRK01458.1 EDW93861.1 GFDF01002803 JAV11281.1 GFDF01002854 JAV11230.1 GBYB01012884 JAG82651.1 CM002912 KMY97239.1 AE014296 BT100048 AAG22234.1 ACX61589.1 CH916366 EDV95630.1 KMY97238.1 ABW08446.1 KMY97232.1 CM000363 EDX09012.1 KMY97230.1 KMY97233.1 CH954178 KQS43231.1 BT125980 AAN11524.1 AAS64952.1 ADX35959.1 AAN11526.1 KQS43226.1 KMY97237.1 AY061183 KQS43227.1 AAF47713.3 CH480817 EDW50266.1 EDV50368.1 CH902618 KPU78244.1 KPU78243.1 BT120205 ADB91453.1 KPU78242.1 EDV39844.1 OUUW01000002 SPP77411.1 SPP77412.1 CH921324 EDV90450.1 CH477190 EAT48725.1 SPP77410.1 CH379070 KRT08719.1 KRT08722.1 CP012525 ALC43112.1 KRT08723.1 EAL30232.2 KRT08721.1 CH964161 EDW79975.2 KRT08720.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000002358
UP000235965
+ More
UP000078540 UP000053097 UP000279307 UP000078542 UP000007755 UP000005205 UP000000311 UP000008237 UP000053825 UP000075809 UP000078541 UP000005203 UP000078492 UP000095300 UP000053105 UP000037069 UP000076502 UP000242457 UP000002282 UP000000803 UP000001070 UP000095301 UP000000304 UP000008711 UP000001292 UP000007801 UP000192221 UP000268350 UP000008820 UP000001819 UP000092553 UP000007798
UP000078540 UP000053097 UP000279307 UP000078542 UP000007755 UP000005205 UP000000311 UP000008237 UP000053825 UP000075809 UP000078541 UP000005203 UP000078492 UP000095300 UP000053105 UP000037069 UP000076502 UP000242457 UP000002282 UP000000803 UP000001070 UP000095301 UP000000304 UP000008711 UP000001292 UP000007801 UP000192221 UP000268350 UP000008820 UP000001819 UP000092553 UP000007798
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9ITL9
A0A2H1V790
A0A2A4K171
A0A2A4JZI1
A0A1E1WSE2
A0A194PRW4
+ More
A0A212EMS4 A0A1E1WHJ0 A0A2A4K041 A0A2A4K0H3 A0A1B6C5K5 K7J653 A0A2J7R474 A0A195BX79 A0A026W4B0 A0A195CAS4 A0A2J7R467 F4WJC7 A0A158NE99 E2A0A4 A0A310SAN1 E2B645 A0A0L7R833 A0A151WSG5 A0A195FQU3 A0A088A552 A0A151J0C7 A0A1I8PZ41 A0A1B6K0L5 E9J1J2 A0A1B6IQW3 A0A0M9A5G5 A0A1I8PZ25 A0A0L0CAC0 A0A1Y1L9N6 A0A034V5D0 A0A1B6MBR2 A0A0K8V1P7 A0A0A1XHV3 A0A154P8V5 A0A0K8V0I2 A0A2A3E6M8 T1P9Y8 A0A0R1DWT5 A0A0R1E1H7 B4PCV7 A0A1L8DXV8 A0A1L8DXP7 A0A0C9R933 A0A0J9RPD0 Q9I7T8 B4IZP5 A0A0J9RMW2 A8JNJ2 A0A0J9RNB8 A0A1I8NAT2 B4QP17 A0A0J9RMV5 A0A0Q5U287 Q7KV83 Q8IRE7 A0A0Q5UH76 A0A1I8NAU6 A0A0J9RNC2 Q95RR8 A0A0Q5U300 Q9VZV3 A0A1I8NAU1 A0A1I8NAT9 B4HTJ5 B3NF26 A0A0N8P0W3 A0A0P8ZUK3 D3DMN9 A0A0P9C3J6 B3M7E6 A0A1W4W1Q9 A0A1W4VNY6 A0A1W4VNP1 A0A1W4W118 A0A3B0JXG1 A0A3B0JBJ9 B4K3A2 Q17PM7 A0A3B0JPK7 A0A0R3P8J3 A0A0R3P6E2 A0A0M4EXP0 A0A0R3P8Z1 Q29E31 A0A0R3P6D7 B4N6I6 A0A0R3P962
A0A212EMS4 A0A1E1WHJ0 A0A2A4K041 A0A2A4K0H3 A0A1B6C5K5 K7J653 A0A2J7R474 A0A195BX79 A0A026W4B0 A0A195CAS4 A0A2J7R467 F4WJC7 A0A158NE99 E2A0A4 A0A310SAN1 E2B645 A0A0L7R833 A0A151WSG5 A0A195FQU3 A0A088A552 A0A151J0C7 A0A1I8PZ41 A0A1B6K0L5 E9J1J2 A0A1B6IQW3 A0A0M9A5G5 A0A1I8PZ25 A0A0L0CAC0 A0A1Y1L9N6 A0A034V5D0 A0A1B6MBR2 A0A0K8V1P7 A0A0A1XHV3 A0A154P8V5 A0A0K8V0I2 A0A2A3E6M8 T1P9Y8 A0A0R1DWT5 A0A0R1E1H7 B4PCV7 A0A1L8DXV8 A0A1L8DXP7 A0A0C9R933 A0A0J9RPD0 Q9I7T8 B4IZP5 A0A0J9RMW2 A8JNJ2 A0A0J9RNB8 A0A1I8NAT2 B4QP17 A0A0J9RMV5 A0A0Q5U287 Q7KV83 Q8IRE7 A0A0Q5UH76 A0A1I8NAU6 A0A0J9RNC2 Q95RR8 A0A0Q5U300 Q9VZV3 A0A1I8NAU1 A0A1I8NAT9 B4HTJ5 B3NF26 A0A0N8P0W3 A0A0P8ZUK3 D3DMN9 A0A0P9C3J6 B3M7E6 A0A1W4W1Q9 A0A1W4VNY6 A0A1W4VNP1 A0A1W4W118 A0A3B0JXG1 A0A3B0JBJ9 B4K3A2 Q17PM7 A0A3B0JPK7 A0A0R3P8J3 A0A0R3P6E2 A0A0M4EXP0 A0A0R3P8Z1 Q29E31 A0A0R3P6D7 B4N6I6 A0A0R3P962
PDB
4BWP
E-value=2.52808e-105,
Score=979
Ontologies
GO
PANTHER
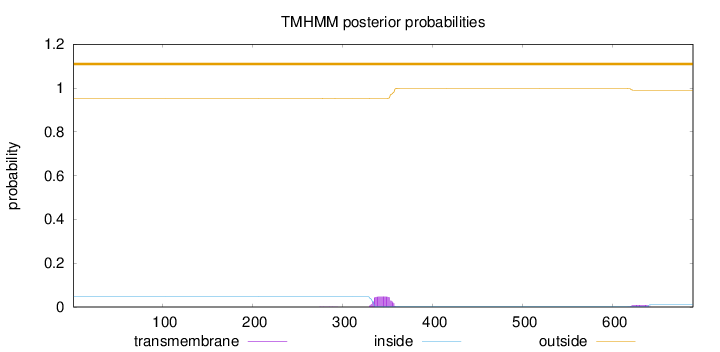
Topology
Subcellular location
Length:
689
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21627
Exp number, first 60 AAs:
0.00025
Total prob of N-in:
0.04735
outside
1 - 689
Population Genetic Test Statistics
Pi
16.273267
Theta
2.383071
Tajima's D
-2.391949
CLR
0.392864
CSRT
0
Interpretation
Uncertain
