Gene
KWMTBOMO00179
Pre Gene Modal
BGIBMGA000600
Annotation
PREDICTED:_phospholipase_C_gamma_isoform_X2_[Bombyx_mori]
Full name
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma
+ More
Phosphoinositide phospholipase C
Phosphoinositide phospholipase C
Location in the cell
Nuclear Reliability : 3.333
Sequence
CDS
ATGAAGTCGGTCTGCTTCAACGGGGCCAAAGATAGGCTCATACACGAGGCAGACCAGCTTATCTCCTACATAGAAAGAGGCTCTACCGTTTTAAAATTCTTCCCTCGTAAGAGACCCGAAAGGCGTGCACTTTGCGTGAGAAGAGAGACCCATCAAGTTCTTTGGTCTCGAATCAATGCCCCTCAGAGACAGGGCTACGAAGGGGCTCTCGATATCCGTGATGTCAAAGAAGTTCGCACTGGAAAATCATCCAAAGACTTTGAGCGTTGGCCTGAAGAAACCAAACGTCTTGAAAGTTCAAAATGCTTTACCATTTATTATGGAACTGAATTTAAACTTCGCTGTGCATCATTTGTTGCTCAAACAGATAAAGAGTGTGAAGCTTGGGTGAAAGGTGTACTGTATCTAATAGCAGAAGCTGTCAGTGCATCATATCCTCTACAGATTGAAAGATGGCTCCGGAAAGAATTCTATTCTATTGAAAACTCCCATGAAAAAATAACATTAAAAGAGGTCAAAGCTTTTCTACCAAAAATTAATTGTAAAATTTCAACAAGTAAGCTTCGAGATGTATTCCAAGATGTTGACACTGAAAAAAGAGGGGAGATTGGCTTTGATGATTTTGCTGTTCTTTATCAGAAGCTAATTTTTGATGAGAATAATGTTCAGGACATATTTGATAAATACTCAGTCTATTGTTCTAATGGTACTACTATTACTCTTAGAGAGTTTCAAAATTTCCTTAGAGATGAACAGCATGATAACCTTGGTGATGATGAAGTACAAGCCTCACAGTTCATCCGTGATTACTTAAGAGATCCACAAAGAGACATCTATGAACCTTATTTCACTTTAAGCGAGTTTGTCGAGATGCTGTACTCCAAGCAGAATTCCATCTATGACAGTCAGTATGATAAGGTTACACAAGATATGACCAGGCCTTTGTCACACTATTGGATATCTTCTTCTCATAACACGTATCTGACAGGTGACCAGTTTTCAAGTGAATCATCACTGGAGGCCTACGTGCGGTGCTTGAGGTCAGGCTGTCGTTGTATTGAGCTAGATTGTTGGGATGGTCCTGACGGCACACCTTTTATTTATCACGGGCACACACTGACAACGAAAATAAGGTTTATGGATGTACTGCGAACAATAAAAGAACATGCATTCATCACATCTGAATATCCATTAATTTTGTCTATCGAAGACAACTGCTCACTCCCACAGCAAAGACGAATGGCGAGCGCATTTCAAGACGTATTCGGTGACCTTCTTCTTGTTCATCCAATGGATAAGAATGAAACATCACTTCCCTCTCCTCATGACTTACGCAGAAAAATAATCCTGAAACATAAAAAATTACCTGAAGGTGCTGAAGAAAGTTCTTTCGCAGTACGTCAAGAAGAAGGAAAAGATTTAGACCTACGAAATTCGATCAAAAATGGTATATTGCATTTGGAAGATCCTGTCGACAAAGAATGGAAGCCTCACGCCTTCGTGCTCACCGAGAACAAACTTTACTATACAGAGAGCTACAACAGTCAAGAAGAAACCGACACTGAAAGTGACGGAGACTCAGATGCGGAAAACGCTATTGTACCACAAGATGAATTACATTTCGCAGAATGTTGGTTCCACGGGAAGCTGGCAGGGAATCGACAAGAAGCCGAAGATTTGCTGAGAGCCCACGCCCATCTTGGCGATGGAACCTTTCTGGTGAGGGAGAGCGTTACTTTTGTCGGTGACTACTGTCTATCATTCTGGCGGCAAGGGAAAGTAAATCACTGCCGCATCAAGCTCAAACAGGAACGGGGGGTCACCAAGTTCTATCTGATAGATAGTATGTGTTTCGACAGCCTGTACAACCTCATCACCCATTATAGGTCACATCCTCTGAGGAGTCAAGAGTTCCTGATAACGTTAAAGGAGCCAGTGCCTCAGCCTAACAAGCATGAAGGCAAGGAATGGTTCCACGCGCACTGCACGCGTACTCAAGCCGAAGAATTACTGCGGAAGACCAACACCGACGGCGCTTTCCTAGTACGCCCCAGCGAAAAGGAACAAGGGAGCTTCGCAATTAGTTTCAGGACTGAGAAGGAGATAAAGCATTGCAGGGTTAAACAAGAGGGTCGCCTGTACACAATCGCGAAGAAACCGTCAGAAGACTTGTCTCTGTAA
Protein
MKSVCFNGAKDRLIHEADQLISYIERGSTVLKFFPRKRPERRALCVRRETHQVLWSRINAPQRQGYEGALDIRDVKEVRTGKSSKDFERWPEETKRLESSKCFTIYYGTEFKLRCASFVAQTDKECEAWVKGVLYLIAEAVSASYPLQIERWLRKEFYSIENSHEKITLKEVKAFLPKINCKISTSKLRDVFQDVDTEKRGEIGFDDFAVLYQKLIFDENNVQDIFDKYSVYCSNGTTITLREFQNFLRDEQHDNLGDDEVQASQFIRDYLRDPQRDIYEPYFTLSEFVEMLYSKQNSIYDSQYDKVTQDMTRPLSHYWISSSHNTYLTGDQFSSESSLEAYVRCLRSGCRCIELDCWDGPDGTPFIYHGHTLTTKIRFMDVLRTIKEHAFITSEYPLILSIEDNCSLPQQRRMASAFQDVFGDLLLVHPMDKNETSLPSPHDLRRKIILKHKKLPEGAEESSFAVRQEEGKDLDLRNSIKNGILHLEDPVDKEWKPHAFVLTENKLYYTESYNSQEETDTESDGDSDAENAIVPQDELHFAECWFHGKLAGNRQEAEDLLRAHAHLGDGTFLVRESVTFVGDYCLSFWRQGKVNHCRIKLKQERGVTKFYLIDSMCFDSLYNLITHYRSHPLRSQEFLITLKEPVPQPNKHEGKEWFHAHCTRTQAEELLRKTNTDGAFLVRPSEKEQGSFAISFRTEKEIKHCRVKQEGRLYTIAKKPSEDLSL
Summary
Description
Mediates the production of the second messenger molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). Plays an important role in the regulation of intracellular signaling cascades.
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = 1D-myo-inositol 1,4,5-trisphosphate + a 1,2-diacyl-sn-glycerol + H(+)
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9ITM0
D2XZ03
A0A2A4K1A7
A0A060BIC2
A0A2H1V5J9
A0A194QW69
+ More
A0A212EMM4 A0A194PRV4 A0A0N1IN36 A0A2J7QPE5 A0A067R4V5 A0A0N0U461 A0A151JQT3 A0A0L7QR48 A0A158NTI1 A0A195C3N1 A0A151HZR5 A0A151X826 A0A026VXP9 A0A3L8D6S6 A0A195FTF0 A0A232F3Z6 A0A2A3EAE0 A0A1B6HJU2 A0A1Y1KG69 A0A1W4XCL1 A0A154P0V9 A0A224X547 A0A023F3P4 A0A310SPU7 A0A0A9X474 A0A139W9D8 D7ELR5 A0A087ZXW1 B0WL59 A0A1S4F7L8 Q17CT3 A0A1Q3F8M6 A0A1Q3F8K4 A0A084VHS7 A0A182QRY2 A0A2R5L918 A0A182RW64 A0A182VF99 A0A182JGV8 A0A182N2B2 A0A131XWM3 A0A182K268 A0A182WDJ1 A0A182KL33 Q7QHE3 A0A182FKL3 A0A182M1X0 A0A182NZM7 A0A182YB20 W5JCJ5 A0A2M4A9Q3 A0A2M4BA71 U4UD37 N6UAF3 B7P4X9 T1IHK2 A0A336LNZ2 A0A182HWB9 A0A182X5M9 A0A224X5R9 A0A1J1IE99 A0A1B0GIU0 A0A1L8DYI1 A0A2M4BAP5 A0A2M4BAQ3 L7MKR7 A0A0P6GX85 A0A0P5Y732 A0A0N8BB53 A0A0P5K5M3 A0A0P6GQA6 A0A182HCQ0 A0A0P5HQ05 A0A1I8NY27 A0A0P5H5I7 A0A0A1WF15 A0A1D2NCI0 W8BW99 A0A1B0FA29 A0A1B0API1 A0A0L0BNN5 A0A1A9ULC6 A0A1A9XWX6 A0A1I8M0T8 A0A1B0AE86 A0A1A9W621 A0A0K8VME4 A0A034WI99 A0A2T7NHX7 T1PIV1 Q86RA6 B4JKL8 A0A2C9JPF0 A2IB48
A0A212EMM4 A0A194PRV4 A0A0N1IN36 A0A2J7QPE5 A0A067R4V5 A0A0N0U461 A0A151JQT3 A0A0L7QR48 A0A158NTI1 A0A195C3N1 A0A151HZR5 A0A151X826 A0A026VXP9 A0A3L8D6S6 A0A195FTF0 A0A232F3Z6 A0A2A3EAE0 A0A1B6HJU2 A0A1Y1KG69 A0A1W4XCL1 A0A154P0V9 A0A224X547 A0A023F3P4 A0A310SPU7 A0A0A9X474 A0A139W9D8 D7ELR5 A0A087ZXW1 B0WL59 A0A1S4F7L8 Q17CT3 A0A1Q3F8M6 A0A1Q3F8K4 A0A084VHS7 A0A182QRY2 A0A2R5L918 A0A182RW64 A0A182VF99 A0A182JGV8 A0A182N2B2 A0A131XWM3 A0A182K268 A0A182WDJ1 A0A182KL33 Q7QHE3 A0A182FKL3 A0A182M1X0 A0A182NZM7 A0A182YB20 W5JCJ5 A0A2M4A9Q3 A0A2M4BA71 U4UD37 N6UAF3 B7P4X9 T1IHK2 A0A336LNZ2 A0A182HWB9 A0A182X5M9 A0A224X5R9 A0A1J1IE99 A0A1B0GIU0 A0A1L8DYI1 A0A2M4BAP5 A0A2M4BAQ3 L7MKR7 A0A0P6GX85 A0A0P5Y732 A0A0N8BB53 A0A0P5K5M3 A0A0P6GQA6 A0A182HCQ0 A0A0P5HQ05 A0A1I8NY27 A0A0P5H5I7 A0A0A1WF15 A0A1D2NCI0 W8BW99 A0A1B0FA29 A0A1B0API1 A0A0L0BNN5 A0A1A9ULC6 A0A1A9XWX6 A0A1I8M0T8 A0A1B0AE86 A0A1A9W621 A0A0K8VME4 A0A034WI99 A0A2T7NHX7 T1PIV1 Q86RA6 B4JKL8 A0A2C9JPF0 A2IB48
EC Number
3.1.4.11
Pubmed
19121390
26354079
22118469
24845553
21347285
24508170
+ More
30249741 28648823 28004739 25474469 25401762 26823975 18362917 19820115 17510324 24438588 20966253 12364791 14747013 17210077 25244985 20920257 23761445 23537049 25576852 26483478 25830018 27289101 24495485 26108605 25315136 25348373 12807765 17994087 18057021 15562597 18951372
30249741 28648823 28004739 25474469 25401762 26823975 18362917 19820115 17510324 24438588 20966253 12364791 14747013 17210077 25244985 20920257 23761445 23537049 25576852 26483478 25830018 27289101 24495485 26108605 25315136 25348373 12807765 17994087 18057021 15562597 18951372
EMBL
BABH01006728
BABH01006729
BABH01006730
BABH01006731
GU266212
ADB25315.1
+ More
NWSH01000326 PCG77422.1 KF800735 AIA77273.1 ODYU01000800 SOQ36125.1 KQ461108 KPJ09210.1 AGBW02013811 OWR42742.1 KQ459594 KPI96176.1 KQ459285 KPJ02044.1 NEVH01012091 PNF30464.1 KK852891 KDR14243.1 KQ435834 KOX71582.1 KQ978627 KYN29563.1 KQ414784 KOC61102.1 ADTU01002746 ADTU01002747 KQ978344 KYM94788.1 KQ976693 KYM77666.1 KQ982431 KYQ56438.1 KK107796 EZA47604.1 QOIP01000013 RLU15548.1 KQ981268 KYN43890.1 NNAY01001051 OXU25332.1 KZ288308 PBC28697.1 GECU01032726 JAS74980.1 GEZM01084973 JAV60354.1 KQ434796 KZC05565.1 GFTR01008801 JAW07625.1 GBBI01002697 JAC16015.1 KQ761834 OAD56687.1 GBHO01028815 GDHC01018996 JAG14789.1 JAP99632.1 KQ972349 KXZ75916.1 EFA12319.2 DS231980 EDS30232.1 CH477304 EAT44189.1 GFDL01011149 JAV23896.1 GFDL01011186 JAV23859.1 ATLV01013225 ATLV01013226 ATLV01013227 ATLV01013228 KE524847 KFB37521.1 AXCN02001729 GGLE01001829 MBY05955.1 GEFM01005730 GEGO01006352 JAP70066.1 JAR89052.1 AAAB01008816 EAA05135.4 AXCM01007440 ADMH02001908 ETN60549.1 GGFK01004174 MBW37495.1 GGFJ01000788 MBW49929.1 KB632225 ERL90238.1 APGK01036671 KB740941 ENN77611.1 ABJB010334257 ABJB010359695 ABJB011028291 DS637651 EEC01651.1 JH429910 UFQT01000094 SSX19640.1 APCN01001484 GFTR01008591 JAW07835.1 CVRI01000047 CRK97326.1 AJWK01017652 AJWK01017653 AJWK01017654 GFDF01002563 JAV11521.1 GGFJ01000965 MBW50106.1 GGFJ01000966 MBW50107.1 GACK01000489 JAA64545.1 GDIQ01027654 JAN67083.1 GDIP01063285 LRGB01000512 JAM40430.1 KZS18530.1 GDIQ01195035 JAK56690.1 GDIQ01249526 GDIQ01247932 GDIQ01244786 GDIQ01244785 GDIQ01244784 GDIQ01233177 GDIQ01231433 GDIQ01205290 GDIQ01203629 GDIQ01190495 GDIQ01187843 GDIQ01171695 GDIQ01163660 GDIQ01162303 GDIQ01160901 GDIQ01140665 GDIQ01138515 GDIQ01130969 GDIQ01126968 GDIQ01125268 GDIQ01123316 GDIQ01116817 GDIQ01116816 GDIQ01096920 GDIP01131328 GDIQ01084279 GDIQ01063042 GDIQ01046045 GDIQ01044407 GDIQ01033749 GDIQ01032297 GDIQ01030610 JAK61230.1 JAL72386.1 GDIQ01030314 JAN64423.1 JXUM01033613 JXUM01033614 JXUM01033615 JXUM01033616 GDIQ01223973 JAK27752.1 GDIQ01237785 GDIQ01237784 JAK13941.1 GBXI01017227 JAC97064.1 LJIJ01000091 ODN02952.1 GAMC01013111 JAB93444.1 CCAG010004929 JXJN01001447 JRES01001597 KNC21621.1 GDHF01012286 JAI40028.1 GAKP01003676 JAC55276.1 PZQS01000012 PVD20783.1 KA648696 AFP63325.1 AF543827 CH940651 AAN39331.1 EDW65634.1 CH916370 EDW00121.1 EF185302 ABM55782.1
NWSH01000326 PCG77422.1 KF800735 AIA77273.1 ODYU01000800 SOQ36125.1 KQ461108 KPJ09210.1 AGBW02013811 OWR42742.1 KQ459594 KPI96176.1 KQ459285 KPJ02044.1 NEVH01012091 PNF30464.1 KK852891 KDR14243.1 KQ435834 KOX71582.1 KQ978627 KYN29563.1 KQ414784 KOC61102.1 ADTU01002746 ADTU01002747 KQ978344 KYM94788.1 KQ976693 KYM77666.1 KQ982431 KYQ56438.1 KK107796 EZA47604.1 QOIP01000013 RLU15548.1 KQ981268 KYN43890.1 NNAY01001051 OXU25332.1 KZ288308 PBC28697.1 GECU01032726 JAS74980.1 GEZM01084973 JAV60354.1 KQ434796 KZC05565.1 GFTR01008801 JAW07625.1 GBBI01002697 JAC16015.1 KQ761834 OAD56687.1 GBHO01028815 GDHC01018996 JAG14789.1 JAP99632.1 KQ972349 KXZ75916.1 EFA12319.2 DS231980 EDS30232.1 CH477304 EAT44189.1 GFDL01011149 JAV23896.1 GFDL01011186 JAV23859.1 ATLV01013225 ATLV01013226 ATLV01013227 ATLV01013228 KE524847 KFB37521.1 AXCN02001729 GGLE01001829 MBY05955.1 GEFM01005730 GEGO01006352 JAP70066.1 JAR89052.1 AAAB01008816 EAA05135.4 AXCM01007440 ADMH02001908 ETN60549.1 GGFK01004174 MBW37495.1 GGFJ01000788 MBW49929.1 KB632225 ERL90238.1 APGK01036671 KB740941 ENN77611.1 ABJB010334257 ABJB010359695 ABJB011028291 DS637651 EEC01651.1 JH429910 UFQT01000094 SSX19640.1 APCN01001484 GFTR01008591 JAW07835.1 CVRI01000047 CRK97326.1 AJWK01017652 AJWK01017653 AJWK01017654 GFDF01002563 JAV11521.1 GGFJ01000965 MBW50106.1 GGFJ01000966 MBW50107.1 GACK01000489 JAA64545.1 GDIQ01027654 JAN67083.1 GDIP01063285 LRGB01000512 JAM40430.1 KZS18530.1 GDIQ01195035 JAK56690.1 GDIQ01249526 GDIQ01247932 GDIQ01244786 GDIQ01244785 GDIQ01244784 GDIQ01233177 GDIQ01231433 GDIQ01205290 GDIQ01203629 GDIQ01190495 GDIQ01187843 GDIQ01171695 GDIQ01163660 GDIQ01162303 GDIQ01160901 GDIQ01140665 GDIQ01138515 GDIQ01130969 GDIQ01126968 GDIQ01125268 GDIQ01123316 GDIQ01116817 GDIQ01116816 GDIQ01096920 GDIP01131328 GDIQ01084279 GDIQ01063042 GDIQ01046045 GDIQ01044407 GDIQ01033749 GDIQ01032297 GDIQ01030610 JAK61230.1 JAL72386.1 GDIQ01030314 JAN64423.1 JXUM01033613 JXUM01033614 JXUM01033615 JXUM01033616 GDIQ01223973 JAK27752.1 GDIQ01237785 GDIQ01237784 JAK13941.1 GBXI01017227 JAC97064.1 LJIJ01000091 ODN02952.1 GAMC01013111 JAB93444.1 CCAG010004929 JXJN01001447 JRES01001597 KNC21621.1 GDHF01012286 JAI40028.1 GAKP01003676 JAC55276.1 PZQS01000012 PVD20783.1 KA648696 AFP63325.1 AF543827 CH940651 AAN39331.1 EDW65634.1 CH916370 EDW00121.1 EF185302 ABM55782.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000235965
+ More
UP000027135 UP000053105 UP000078492 UP000053825 UP000005205 UP000078542 UP000078540 UP000075809 UP000053097 UP000279307 UP000078541 UP000215335 UP000242457 UP000192223 UP000076502 UP000007266 UP000005203 UP000002320 UP000008820 UP000030765 UP000075886 UP000075900 UP000075903 UP000075880 UP000075884 UP000075881 UP000075920 UP000075882 UP000007062 UP000069272 UP000075883 UP000075885 UP000076408 UP000000673 UP000030742 UP000019118 UP000001555 UP000075840 UP000076407 UP000183832 UP000092461 UP000076858 UP000069940 UP000095300 UP000094527 UP000092444 UP000092460 UP000037069 UP000078200 UP000092443 UP000095301 UP000092445 UP000091820 UP000245119 UP000008792 UP000001070 UP000076420
UP000027135 UP000053105 UP000078492 UP000053825 UP000005205 UP000078542 UP000078540 UP000075809 UP000053097 UP000279307 UP000078541 UP000215335 UP000242457 UP000192223 UP000076502 UP000007266 UP000005203 UP000002320 UP000008820 UP000030765 UP000075886 UP000075900 UP000075903 UP000075880 UP000075884 UP000075881 UP000075920 UP000075882 UP000007062 UP000069272 UP000075883 UP000075885 UP000076408 UP000000673 UP000030742 UP000019118 UP000001555 UP000075840 UP000076407 UP000183832 UP000092461 UP000076858 UP000069940 UP000095300 UP000094527 UP000092444 UP000092460 UP000037069 UP000078200 UP000092443 UP000095301 UP000092445 UP000091820 UP000245119 UP000008792 UP000001070 UP000076420
Pfam
Interpro
IPR035024
PLC-gamma_N-SH2
+ More
IPR016279 PLC-gamma
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR001192 PI-PLC_fam
IPR001711 PLipase_C_Pinositol-sp_Y
IPR036860 SH2_dom_sf
IPR035892 C2_domain_sf
IPR000980 SH2
IPR002048 EF_hand_dom
IPR000008 C2_dom
IPR011992 EF-hand-dom_pair
IPR035023 PLC-gamma_C-SH2
IPR011993 PH-like_dom_sf
IPR010721 DUF1295
IPR001104 3-oxo-5_a-steroid_4-DH_C
IPR015359 PLC_EF-hand-like
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
IPR016279 PLC-gamma
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR001849 PH_domain
IPR001192 PI-PLC_fam
IPR001711 PLipase_C_Pinositol-sp_Y
IPR036860 SH2_dom_sf
IPR035892 C2_domain_sf
IPR000980 SH2
IPR002048 EF_hand_dom
IPR000008 C2_dom
IPR011992 EF-hand-dom_pair
IPR035023 PLC-gamma_C-SH2
IPR011993 PH-like_dom_sf
IPR010721 DUF1295
IPR001104 3-oxo-5_a-steroid_4-DH_C
IPR015359 PLC_EF-hand-like
IPR027417 P-loop_NTPase
IPR036961 Kinesin_motor_dom_sf
IPR001752 Kinesin_motor_dom
SUPFAM
ProteinModelPortal
H9ITM0
D2XZ03
A0A2A4K1A7
A0A060BIC2
A0A2H1V5J9
A0A194QW69
+ More
A0A212EMM4 A0A194PRV4 A0A0N1IN36 A0A2J7QPE5 A0A067R4V5 A0A0N0U461 A0A151JQT3 A0A0L7QR48 A0A158NTI1 A0A195C3N1 A0A151HZR5 A0A151X826 A0A026VXP9 A0A3L8D6S6 A0A195FTF0 A0A232F3Z6 A0A2A3EAE0 A0A1B6HJU2 A0A1Y1KG69 A0A1W4XCL1 A0A154P0V9 A0A224X547 A0A023F3P4 A0A310SPU7 A0A0A9X474 A0A139W9D8 D7ELR5 A0A087ZXW1 B0WL59 A0A1S4F7L8 Q17CT3 A0A1Q3F8M6 A0A1Q3F8K4 A0A084VHS7 A0A182QRY2 A0A2R5L918 A0A182RW64 A0A182VF99 A0A182JGV8 A0A182N2B2 A0A131XWM3 A0A182K268 A0A182WDJ1 A0A182KL33 Q7QHE3 A0A182FKL3 A0A182M1X0 A0A182NZM7 A0A182YB20 W5JCJ5 A0A2M4A9Q3 A0A2M4BA71 U4UD37 N6UAF3 B7P4X9 T1IHK2 A0A336LNZ2 A0A182HWB9 A0A182X5M9 A0A224X5R9 A0A1J1IE99 A0A1B0GIU0 A0A1L8DYI1 A0A2M4BAP5 A0A2M4BAQ3 L7MKR7 A0A0P6GX85 A0A0P5Y732 A0A0N8BB53 A0A0P5K5M3 A0A0P6GQA6 A0A182HCQ0 A0A0P5HQ05 A0A1I8NY27 A0A0P5H5I7 A0A0A1WF15 A0A1D2NCI0 W8BW99 A0A1B0FA29 A0A1B0API1 A0A0L0BNN5 A0A1A9ULC6 A0A1A9XWX6 A0A1I8M0T8 A0A1B0AE86 A0A1A9W621 A0A0K8VME4 A0A034WI99 A0A2T7NHX7 T1PIV1 Q86RA6 B4JKL8 A0A2C9JPF0 A2IB48
A0A212EMM4 A0A194PRV4 A0A0N1IN36 A0A2J7QPE5 A0A067R4V5 A0A0N0U461 A0A151JQT3 A0A0L7QR48 A0A158NTI1 A0A195C3N1 A0A151HZR5 A0A151X826 A0A026VXP9 A0A3L8D6S6 A0A195FTF0 A0A232F3Z6 A0A2A3EAE0 A0A1B6HJU2 A0A1Y1KG69 A0A1W4XCL1 A0A154P0V9 A0A224X547 A0A023F3P4 A0A310SPU7 A0A0A9X474 A0A139W9D8 D7ELR5 A0A087ZXW1 B0WL59 A0A1S4F7L8 Q17CT3 A0A1Q3F8M6 A0A1Q3F8K4 A0A084VHS7 A0A182QRY2 A0A2R5L918 A0A182RW64 A0A182VF99 A0A182JGV8 A0A182N2B2 A0A131XWM3 A0A182K268 A0A182WDJ1 A0A182KL33 Q7QHE3 A0A182FKL3 A0A182M1X0 A0A182NZM7 A0A182YB20 W5JCJ5 A0A2M4A9Q3 A0A2M4BA71 U4UD37 N6UAF3 B7P4X9 T1IHK2 A0A336LNZ2 A0A182HWB9 A0A182X5M9 A0A224X5R9 A0A1J1IE99 A0A1B0GIU0 A0A1L8DYI1 A0A2M4BAP5 A0A2M4BAQ3 L7MKR7 A0A0P6GX85 A0A0P5Y732 A0A0N8BB53 A0A0P5K5M3 A0A0P6GQA6 A0A182HCQ0 A0A0P5HQ05 A0A1I8NY27 A0A0P5H5I7 A0A0A1WF15 A0A1D2NCI0 W8BW99 A0A1B0FA29 A0A1B0API1 A0A0L0BNN5 A0A1A9ULC6 A0A1A9XWX6 A0A1I8M0T8 A0A1B0AE86 A0A1A9W621 A0A0K8VME4 A0A034WI99 A0A2T7NHX7 T1PIV1 Q86RA6 B4JKL8 A0A2C9JPF0 A2IB48
PDB
4EY0
E-value=2.51302e-51,
Score=513
Ontologies
PATHWAY
GO
GO:0035556
GO:0004435
GO:0005509
GO:0009395
GO:0016042
GO:0016021
GO:0007165
GO:0016627
GO:0051209
GO:0048015
GO:0032959
GO:0010634
GO:0005886
GO:0007018
GO:0008017
GO:0003777
GO:0005524
GO:0070373
GO:0045873
GO:0042059
GO:0007298
GO:0032868
GO:0030307
GO:0006629
GO:0008081
GO:0005515
GO:0004713
GO:0004672
GO:0003707
GO:0007169
PANTHER
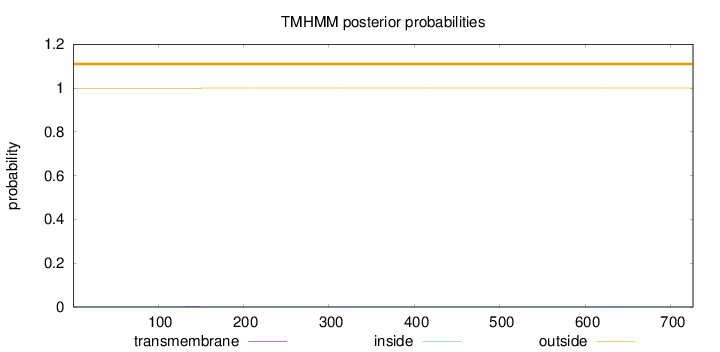
Topology
Length:
726
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05639
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00293
outside
1 - 726
Population Genetic Test Statistics
Pi
0.231142
Theta
1.333538
Tajima's D
-1.898084
CLR
0.838361
CSRT
0.0100994950252487
Interpretation
Possibly Positive selection
