Gene
KWMTBOMO00177
Pre Gene Modal
BGIBMGA000601
Annotation
PREDICTED:_TGF-beta_receptor_type-1_isoform_X2_[Bombyx_mori]
Full name
Serine/threonine-protein kinase receptor
+ More
Receptor protein serine/threonine kinase
Receptor protein serine/threonine kinase
Location in the cell
Nuclear Reliability : 1.317
Sequence
CDS
ATGTGCAATCGAGAGATACTCCAGGAGCTGCATCCGAAGCCGCCAGAGACGAGCCCGACCGCGACGGCATGGCAGGTTGTGCCTTGGATCATGGGACTTGTTGTATTCGCCATATGCGTCGCTTTAAGTCTGTGGTGGGCGAAGAAATGGCGAGCCGACGGGAAACGACCGCCACGGCCTTACCCGGATGAGGACGAGACCAAACACATACTTAATACACCCACCACCACTATCAGAGATATGATTGAACTAACGACGTCTGGCTCTGGATCTGGATTACCGCTTCTGGTGCAGCGAAGTATCGCACGCCAGATCCAACTGGTGGACATAATTGGAAAGGGTCGTTTCGGCGAGGTGTGGCGTGGTCGTTGGAGGGGTGAGAACGTCGCGGTTAAGATATTTTCATCTCGCGAGGAATGTTCTTGGTTCCGGGAAGCGGAAATTTACCAGACCGTGATGTTGCGGCATGAAAATATCCTTGGGTTCATTGCGGCTGATAATAAAGATAATGGGACGTGGACTCAGTTATGGTTGATAACTGATTATCATGAAAATGGTTCTCTATTTGATTTCCTTACTGCAAAATCAATCGACTCGAACACGCTCATTAAGATGTCCCTCTCCATCGCAACTGGACTAGCTCATTTGCATATGGACATTGTCGGCACAAAAGGAAAGCCGGCTATAGCTCATCGGGATCTCAAATCGAAAAACATATTGGTGAAAAGCAACTTGTCTTGCGTGATCGGGGATCTCGGGCTTGCCGTGCGCCACAATGTCTCGAACGACTCTGTTGATGTACCGTCGACCAACCGTGTGGGCACCAAGCGGTACATGGCGCCGGAGGTATTAGATGAAACGATGGACACGCGGCAGTTCGATCCTTACAAGCGTTCGGACGTCTATTCATTCGGCCTAGTGCTGTGGGAGATGGCTCGTCGCTGCGGTAACATGCCGGACGATTACCAGCCTCCTTATTACGACTGCGTGCCACCGGATCCGGCGCTCGAAGACATGAGACGCGTTGTATGCACCGAGAAGCGAAGACCGAACGTACCTAATAGGTGGCACTCTGATCCGGTGCTGTCAGGAATATCAAAGGTAATGAAAGAATGCTGGTACCAAAATCCGGCGGCCAGGTTGACTGCGCTCCGAATCAAGAAAACCCTTGCCAATGTCGGAACAAACGGCGCGGACTACATCAAACTTTGA
Protein
MCNREILQELHPKPPETSPTATAWQVVPWIMGLVVFAICVALSLWWAKKWRADGKRPPRPYPDEDETKHILNTPTTTIRDMIELTTSGSGSGLPLLVQRSIARQIQLVDIIGKGRFGEVWRGRWRGENVAVKIFSSREECSWFREAEIYQTVMLRHENILGFIAADNKDNGTWTQLWLITDYHENGSLFDFLTAKSIDSNTLIKMSLSIATGLAHLHMDIVGTKGKPAIAHRDLKSKNILVKSNLSCVIGDLGLAVRHNVSNDSVDVPSTNRVGTKRYMAPEVLDETMDTRQFDPYKRSDVYSFGLVLWEMARRCGNMPDDYQPPYYDCVPPDPALEDMRRVVCTEKRRPNVPNRWHSDPVLSGISKVMKECWYQNPAARLTALRIKKTLANVGTNGADYIKL
Summary
Catalytic Activity
[receptor-protein]-L-serine + ATP = [receptor-protein]-O-phospho-L-serine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
[receptor-protein]-L-threonine + ATP = [receptor-protein]-O-phospho-L-threonine + ADP + H(+)
Similarity
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family. TGFB receptor subfamily.
Belongs to the protein kinase superfamily.
Belongs to the protein kinase superfamily.
Feature
chain Serine/threonine-protein kinase receptor
Uniprot
H9ITM1
A0A220QT61
A0A194QVZ3
A0A2A4K181
A0A2A4K0I3
A0A2A4K085
+ More
A0A2A4JZI3 A0A2A4K1B7 A0A2A4K052 A0A2A4JZJ2 A0A194Q6G9 A0A212EMN2 A0A2A4K0Z4 A0A2A4JZK3 A0A2A4K0Y3 A0A2A4K0J8 A0A2A4K0K7 A0A2H1V5S4 A0A1Q3G0F9 A0A1B0D486 A0A1W4UY47 A0A1W4UZF9 A0A1W4UKW5 N6W4E6 A0A3B0IZZ5 B4GIN9 A0A1W4UXU3 A0A1I8NCQ2 A0A0R3NNR5 A0A0P8XMK2 B4QGP3 A0A3B0IZP4 A0A0R3NNL0 B4P3F5 A0A1I8PXB6 B4J5D9 A0A0J9R8L6 A0A0R3NNU5 B3N833 A0A0P9ACU6 A0A0R1DTJ7 A0A0Q5WBX1 A0A0J9TWT2 Q28YV2 Q23975 A0A0Q5WKQ1 A0A1I8NCP3 A0A1I8PXB2 A0A0J9R8G5 A0A0Q9XHD7 A1Z7L9 A0A0R1DTF7 A0A1I8NCP7 B3MI97 Q7YU60 A0A3B0JL14 Q17KD7 A0A1I8PXG0 A0A0A1WZ77 A0A0Q9X2Q0 A0A1I8PXC9 A1Z7L8 B4N626 A0A182H9M3 A0A034WMN3 A0A0A1WW58 B4HSC7 A0A1B0ETQ7 A0A0K8WBI6 U5EYZ8 A0A0K8U047 A0A0A1WVW6 B4KLN9 A0A0Q9X7D9 A0A1L8DJT4 A0A1L8DK04 W8AUF0 A0A1A9W2G1 A0A1L8DJN7 A0A1L8DJM3 A0A1A9UZD4 W8B4V7 A0A1A9YEV6 A0A1B0BKH0 A0A0L0CMN7 A0A1B0GA96 A0A084W4S3 B4LLJ2 A0A0Q9WDW8 A0A0Q9W1Y9 A0A1Y9IVJ1 A0A182QTN6 A0A182LTG0 A0A182NU34 A0A1Y9IV94 A0A0Q9W1Y0 A0A1B6FNR6 A0A1B6JF99 A0A2M4ANU6 A0A151IUW5
A0A2A4JZI3 A0A2A4K1B7 A0A2A4K052 A0A2A4JZJ2 A0A194Q6G9 A0A212EMN2 A0A2A4K0Z4 A0A2A4JZK3 A0A2A4K0Y3 A0A2A4K0J8 A0A2A4K0K7 A0A2H1V5S4 A0A1Q3G0F9 A0A1B0D486 A0A1W4UY47 A0A1W4UZF9 A0A1W4UKW5 N6W4E6 A0A3B0IZZ5 B4GIN9 A0A1W4UXU3 A0A1I8NCQ2 A0A0R3NNR5 A0A0P8XMK2 B4QGP3 A0A3B0IZP4 A0A0R3NNL0 B4P3F5 A0A1I8PXB6 B4J5D9 A0A0J9R8L6 A0A0R3NNU5 B3N833 A0A0P9ACU6 A0A0R1DTJ7 A0A0Q5WBX1 A0A0J9TWT2 Q28YV2 Q23975 A0A0Q5WKQ1 A0A1I8NCP3 A0A1I8PXB2 A0A0J9R8G5 A0A0Q9XHD7 A1Z7L9 A0A0R1DTF7 A0A1I8NCP7 B3MI97 Q7YU60 A0A3B0JL14 Q17KD7 A0A1I8PXG0 A0A0A1WZ77 A0A0Q9X2Q0 A0A1I8PXC9 A1Z7L8 B4N626 A0A182H9M3 A0A034WMN3 A0A0A1WW58 B4HSC7 A0A1B0ETQ7 A0A0K8WBI6 U5EYZ8 A0A0K8U047 A0A0A1WVW6 B4KLN9 A0A0Q9X7D9 A0A1L8DJT4 A0A1L8DK04 W8AUF0 A0A1A9W2G1 A0A1L8DJN7 A0A1L8DJM3 A0A1A9UZD4 W8B4V7 A0A1A9YEV6 A0A1B0BKH0 A0A0L0CMN7 A0A1B0GA96 A0A084W4S3 B4LLJ2 A0A0Q9WDW8 A0A0Q9W1Y9 A0A1Y9IVJ1 A0A182QTN6 A0A182LTG0 A0A182NU34 A0A1Y9IV94 A0A0Q9W1Y0 A0A1B6FNR6 A0A1B6JF99 A0A2M4ANU6 A0A151IUW5
EC Number
2.7.11.30
Pubmed
EMBL
BABH01006725
BABH01006726
BABH01006727
KY328721
ASK12089.1
KQ461108
+ More
KPJ09135.1 NWSH01000326 PCG77430.1 PCG77428.1 PCG77426.1 PCG77425.1 PCG77432.1 PCG77429.1 PCG77431.1 KQ459463 KPJ00600.1 AGBW02013811 OWR42744.1 PCG77433.1 PCG77427.1 PCG77423.1 PCG77424.1 PCG77434.1 ODYU01000800 SOQ36126.1 GFDL01001768 JAV33277.1 AJVK01011278 CM000071 ENO01712.1 OUUW01000001 SPP73925.1 CH479183 EDW36359.1 KRT02631.1 CH902619 KPU75898.1 CM000362 CM002911 EDX06268.1 KMY92389.1 SPP73924.1 KRT02632.1 CM000157 EDW89428.1 CH916367 EDW01781.1 KMY92391.1 KRT02633.1 CH954177 EDV59446.1 KPU75897.1 KRJ98433.1 KQS70828.1 KMY92390.1 EAL25863.3 U04692 AAA18959.1 KQS70827.1 KMY92392.1 CH933808 KRG04222.1 AE013599 AAF59011.1 KRJ98432.1 EDV35942.2 BT009976 AAQ22445.1 ACZ94371.1 SPP73926.1 CH477225 EAT47154.1 GBXI01009943 JAD04349.1 CH964154 KRF99165.1 AAM71094.1 EDW79815.2 JXUM01030571 JXUM01030572 JXUM01030573 KQ560889 KXJ80467.1 GAKP01003914 JAC55038.1 GBXI01011185 JAD03107.1 CH480816 EDW47022.1 AJWK01011290 GDHF01003793 JAI48521.1 GANO01001696 JAB58175.1 GDHF01032624 JAI19690.1 GBXI01011719 JAD02573.1 EDW08675.2 KRG04223.1 GFDF01007403 JAV06681.1 GFDF01007404 JAV06680.1 GAMC01018222 JAB88333.1 GFDF01007406 JAV06678.1 GFDF01007405 JAV06679.1 GAMC01018224 JAB88331.1 JXJN01015940 JRES01000182 KNC33546.1 CCAG010018535 ATLV01020378 ATLV01020379 ATLV01020380 KE525299 KFB45217.1 CH940648 EDW59897.1 KRF79147.1 KRF79145.1 AXCN02000776 AXCM01003318 KRF79146.1 GECZ01017970 JAS51799.1 GECU01009841 JAS97865.1 GGFK01009106 MBW42427.1 KQ980945 KYN11252.1
KPJ09135.1 NWSH01000326 PCG77430.1 PCG77428.1 PCG77426.1 PCG77425.1 PCG77432.1 PCG77429.1 PCG77431.1 KQ459463 KPJ00600.1 AGBW02013811 OWR42744.1 PCG77433.1 PCG77427.1 PCG77423.1 PCG77424.1 PCG77434.1 ODYU01000800 SOQ36126.1 GFDL01001768 JAV33277.1 AJVK01011278 CM000071 ENO01712.1 OUUW01000001 SPP73925.1 CH479183 EDW36359.1 KRT02631.1 CH902619 KPU75898.1 CM000362 CM002911 EDX06268.1 KMY92389.1 SPP73924.1 KRT02632.1 CM000157 EDW89428.1 CH916367 EDW01781.1 KMY92391.1 KRT02633.1 CH954177 EDV59446.1 KPU75897.1 KRJ98433.1 KQS70828.1 KMY92390.1 EAL25863.3 U04692 AAA18959.1 KQS70827.1 KMY92392.1 CH933808 KRG04222.1 AE013599 AAF59011.1 KRJ98432.1 EDV35942.2 BT009976 AAQ22445.1 ACZ94371.1 SPP73926.1 CH477225 EAT47154.1 GBXI01009943 JAD04349.1 CH964154 KRF99165.1 AAM71094.1 EDW79815.2 JXUM01030571 JXUM01030572 JXUM01030573 KQ560889 KXJ80467.1 GAKP01003914 JAC55038.1 GBXI01011185 JAD03107.1 CH480816 EDW47022.1 AJWK01011290 GDHF01003793 JAI48521.1 GANO01001696 JAB58175.1 GDHF01032624 JAI19690.1 GBXI01011719 JAD02573.1 EDW08675.2 KRG04223.1 GFDF01007403 JAV06681.1 GFDF01007404 JAV06680.1 GAMC01018222 JAB88333.1 GFDF01007406 JAV06678.1 GFDF01007405 JAV06679.1 GAMC01018224 JAB88331.1 JXJN01015940 JRES01000182 KNC33546.1 CCAG010018535 ATLV01020378 ATLV01020379 ATLV01020380 KE525299 KFB45217.1 CH940648 EDW59897.1 KRF79147.1 KRF79145.1 AXCN02000776 AXCM01003318 KRF79146.1 GECZ01017970 JAS51799.1 GECU01009841 JAS97865.1 GGFK01009106 MBW42427.1 KQ980945 KYN11252.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000007151
UP000092462
+ More
UP000192221 UP000001819 UP000268350 UP000008744 UP000095301 UP000007801 UP000000304 UP000002282 UP000095300 UP000001070 UP000008711 UP000009192 UP000000803 UP000008820 UP000007798 UP000069940 UP000249989 UP000001292 UP000092461 UP000091820 UP000078200 UP000092443 UP000092460 UP000037069 UP000092444 UP000030765 UP000008792 UP000075920 UP000075886 UP000075883 UP000075884 UP000078492
UP000192221 UP000001819 UP000268350 UP000008744 UP000095301 UP000007801 UP000000304 UP000002282 UP000095300 UP000001070 UP000008711 UP000009192 UP000000803 UP000008820 UP000007798 UP000069940 UP000249989 UP000001292 UP000092461 UP000091820 UP000078200 UP000092443 UP000092460 UP000037069 UP000092444 UP000030765 UP000008792 UP000075920 UP000075886 UP000075883 UP000075884 UP000078492
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
H9ITM1
A0A220QT61
A0A194QVZ3
A0A2A4K181
A0A2A4K0I3
A0A2A4K085
+ More
A0A2A4JZI3 A0A2A4K1B7 A0A2A4K052 A0A2A4JZJ2 A0A194Q6G9 A0A212EMN2 A0A2A4K0Z4 A0A2A4JZK3 A0A2A4K0Y3 A0A2A4K0J8 A0A2A4K0K7 A0A2H1V5S4 A0A1Q3G0F9 A0A1B0D486 A0A1W4UY47 A0A1W4UZF9 A0A1W4UKW5 N6W4E6 A0A3B0IZZ5 B4GIN9 A0A1W4UXU3 A0A1I8NCQ2 A0A0R3NNR5 A0A0P8XMK2 B4QGP3 A0A3B0IZP4 A0A0R3NNL0 B4P3F5 A0A1I8PXB6 B4J5D9 A0A0J9R8L6 A0A0R3NNU5 B3N833 A0A0P9ACU6 A0A0R1DTJ7 A0A0Q5WBX1 A0A0J9TWT2 Q28YV2 Q23975 A0A0Q5WKQ1 A0A1I8NCP3 A0A1I8PXB2 A0A0J9R8G5 A0A0Q9XHD7 A1Z7L9 A0A0R1DTF7 A0A1I8NCP7 B3MI97 Q7YU60 A0A3B0JL14 Q17KD7 A0A1I8PXG0 A0A0A1WZ77 A0A0Q9X2Q0 A0A1I8PXC9 A1Z7L8 B4N626 A0A182H9M3 A0A034WMN3 A0A0A1WW58 B4HSC7 A0A1B0ETQ7 A0A0K8WBI6 U5EYZ8 A0A0K8U047 A0A0A1WVW6 B4KLN9 A0A0Q9X7D9 A0A1L8DJT4 A0A1L8DK04 W8AUF0 A0A1A9W2G1 A0A1L8DJN7 A0A1L8DJM3 A0A1A9UZD4 W8B4V7 A0A1A9YEV6 A0A1B0BKH0 A0A0L0CMN7 A0A1B0GA96 A0A084W4S3 B4LLJ2 A0A0Q9WDW8 A0A0Q9W1Y9 A0A1Y9IVJ1 A0A182QTN6 A0A182LTG0 A0A182NU34 A0A1Y9IV94 A0A0Q9W1Y0 A0A1B6FNR6 A0A1B6JF99 A0A2M4ANU6 A0A151IUW5
A0A2A4JZI3 A0A2A4K1B7 A0A2A4K052 A0A2A4JZJ2 A0A194Q6G9 A0A212EMN2 A0A2A4K0Z4 A0A2A4JZK3 A0A2A4K0Y3 A0A2A4K0J8 A0A2A4K0K7 A0A2H1V5S4 A0A1Q3G0F9 A0A1B0D486 A0A1W4UY47 A0A1W4UZF9 A0A1W4UKW5 N6W4E6 A0A3B0IZZ5 B4GIN9 A0A1W4UXU3 A0A1I8NCQ2 A0A0R3NNR5 A0A0P8XMK2 B4QGP3 A0A3B0IZP4 A0A0R3NNL0 B4P3F5 A0A1I8PXB6 B4J5D9 A0A0J9R8L6 A0A0R3NNU5 B3N833 A0A0P9ACU6 A0A0R1DTJ7 A0A0Q5WBX1 A0A0J9TWT2 Q28YV2 Q23975 A0A0Q5WKQ1 A0A1I8NCP3 A0A1I8PXB2 A0A0J9R8G5 A0A0Q9XHD7 A1Z7L9 A0A0R1DTF7 A0A1I8NCP7 B3MI97 Q7YU60 A0A3B0JL14 Q17KD7 A0A1I8PXG0 A0A0A1WZ77 A0A0Q9X2Q0 A0A1I8PXC9 A1Z7L8 B4N626 A0A182H9M3 A0A034WMN3 A0A0A1WW58 B4HSC7 A0A1B0ETQ7 A0A0K8WBI6 U5EYZ8 A0A0K8U047 A0A0A1WVW6 B4KLN9 A0A0Q9X7D9 A0A1L8DJT4 A0A1L8DK04 W8AUF0 A0A1A9W2G1 A0A1L8DJN7 A0A1L8DJM3 A0A1A9UZD4 W8B4V7 A0A1A9YEV6 A0A1B0BKH0 A0A0L0CMN7 A0A1B0GA96 A0A084W4S3 B4LLJ2 A0A0Q9WDW8 A0A0Q9W1Y9 A0A1Y9IVJ1 A0A182QTN6 A0A182LTG0 A0A182NU34 A0A1Y9IV94 A0A0Q9W1Y0 A0A1B6FNR6 A0A1B6JF99 A0A2M4ANU6 A0A151IUW5
PDB
1PY5
E-value=8.29143e-137,
Score=1248
Ontologies
PATHWAY
GO
GO:0046872
GO:0016021
GO:0005524
GO:0004675
GO:0007179
GO:0043235
GO:0048179
GO:0008340
GO:0007476
GO:0007411
GO:0030513
GO:0007455
GO:0048185
GO:0010862
GO:0016319
GO:0032924
GO:0007405
GO:0009411
GO:0007346
GO:0010507
GO:0046332
GO:0005025
GO:0019838
GO:0005887
GO:0007389
GO:0048813
GO:0007165
GO:0004672
GO:0004703
GO:0016361
GO:0005024
GO:0048666
GO:0006468
GO:0005886
GO:0016020
GO:0004713
GO:0003707
GO:0007169
PANTHER
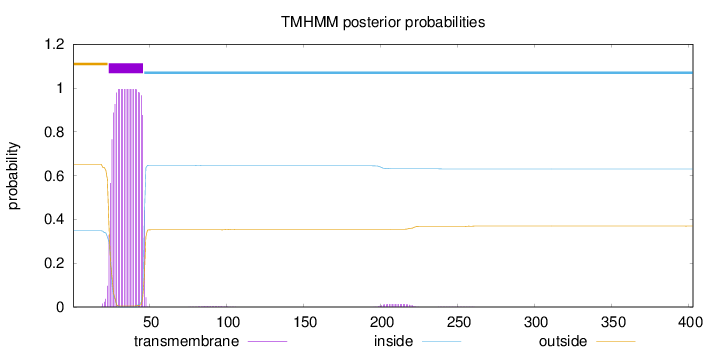
Topology
Subcellular location
Membrane
Length:
403
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.20615
Exp number, first 60 AAs:
21.85738
Total prob of N-in:
0.34797
POSSIBLE N-term signal
sequence
outside
1 - 23
TMhelix
24 - 46
inside
47 - 403
Population Genetic Test Statistics
Pi
0.425956
Theta
2.377599
Tajima's D
-1.939355
CLR
1.623493
CSRT
0.0126993650317484
Interpretation
Possibly Positive selection
