Gene
KWMTBOMO00176
Pre Gene Modal
BGIBMGA000616
Annotation
hypothetical_protein_KGM_15207_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 2.743
Sequence
CDS
ATGTCGGCATCTGAACCTATTGACTTATTTACAGGGAACGGCACGTATAAGTCTATAAACCCCATCATATCAAAAGTCGGTGATTTTACACCTTTTTACATAGCGATAGCTATAAGTTCCGTTATTTTGGGTTTTCTCCTGATTTTAAACATTGTTTGCTGTTGCTCCCGATACTCTGACTATTGGCTCGATAGACACACAGGAAATCGCTGGATTGTTTCCATCTGGTCTACAACCCCTCATAAACAGCCTCCTCTTGACTTGACTGAGTTAAACATTGAAAAATTGCCTATTCAATTTGTGGCACCATACCACAGTGACGAATATCATGAAGTTTTAAGACACGAGTCTGATTTACCCCCATCAATATCCCGTCAGCCTCTCACTTCATCACAAGAGTACTTGGAGTTGCAGAAAAGAGAAAGTGACATTTAG
Protein
MSASEPIDLFTGNGTYKSINPIISKVGDFTPFYIAIAISSVILGFLLILNIVCCCSRYSDYWLDRHTGNRWIVSIWSTTPHKQPPLDLTELNIEKLPIQFVAPYHSDEYHEVLRHESDLPPSISRQPLTSSQEYLELQKRESDI
Summary
Uniprot
H9ITN6
A0A212EMM5
A0A2H1WLP4
A0A2A4K594
I4DKN1
A0A194QUZ0
+ More
S4PZX0 A0A182Y1C1 A0A3F2YWJ7 A0A1L8D932 Q7PNP9 A0A2C9GPQ3 A0A1B0CD90 A0A182Q6I8 A0A026W334 D7EJZ1 B3M569 A0A3B0JBV0 A0A1A9YEH2 A0A1B0B7X0 Q29E76 B4HAT9 A0A2M4CU88 B4MLE0 A0A0K8TMM8 B4KZW2 A0A1A9V479 A0A1A9ZS28 D3TP28 A0A0M4EKI5 A0A088A5C5 B4LFB8 A0A182FG97 V9IGQ3 A0A0L7R827 A0A2M4AQQ3 A0A2M3Z5U4 U5ELL6 A0A232EVD5 B4PK58 K7J651 E1JIE5 A0A182J000 A0A310SET5 B3NFF3 B4HV47 B4QKC3 Q9VS49 B4J371 W8BTA3 E0VZQ8 E2B647 A0A0J7L6C2 A0A2M4C2M7 A0A0K8TYQ0 A0A034V9N7 A0A154P932 T1DFS2 A0A2A3E537 A0A336LSN1 A0A0A1XB99 A0A0T6B7Z6 A0A0L0BXD7 A0A2M4C336 A0A1I8MM87 A0A1Q3FBI2 A0A195CAL2 A0A195BX83 A0A158NEA1 E2A0A7 F4WJC5 A0A069DP63 A0A2P8ZN15 A0A0P4W0U6 A0A1I8Q4L9 B0WIK6 A0A0V0GC25 A0A2M4CU42 A0A0M9A5G3 A0A146M515 A0A2R7WA26 A0A195FKU4
S4PZX0 A0A182Y1C1 A0A3F2YWJ7 A0A1L8D932 Q7PNP9 A0A2C9GPQ3 A0A1B0CD90 A0A182Q6I8 A0A026W334 D7EJZ1 B3M569 A0A3B0JBV0 A0A1A9YEH2 A0A1B0B7X0 Q29E76 B4HAT9 A0A2M4CU88 B4MLE0 A0A0K8TMM8 B4KZW2 A0A1A9V479 A0A1A9ZS28 D3TP28 A0A0M4EKI5 A0A088A5C5 B4LFB8 A0A182FG97 V9IGQ3 A0A0L7R827 A0A2M4AQQ3 A0A2M3Z5U4 U5ELL6 A0A232EVD5 B4PK58 K7J651 E1JIE5 A0A182J000 A0A310SET5 B3NFF3 B4HV47 B4QKC3 Q9VS49 B4J371 W8BTA3 E0VZQ8 E2B647 A0A0J7L6C2 A0A2M4C2M7 A0A0K8TYQ0 A0A034V9N7 A0A154P932 T1DFS2 A0A2A3E537 A0A336LSN1 A0A0A1XB99 A0A0T6B7Z6 A0A0L0BXD7 A0A2M4C336 A0A1I8MM87 A0A1Q3FBI2 A0A195CAL2 A0A195BX83 A0A158NEA1 E2A0A7 F4WJC5 A0A069DP63 A0A2P8ZN15 A0A0P4W0U6 A0A1I8Q4L9 B0WIK6 A0A0V0GC25 A0A2M4CU42 A0A0M9A5G3 A0A146M515 A0A2R7WA26 A0A195FKU4
Pubmed
19121390
22118469
22651552
26354079
23622113
25244985
+ More
12364791 14747013 17210077 24508170 30249741 18362917 19820115 17994087 15632085 26369729 20353571 18057021 28648823 17550304 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356 24495485 20566863 20798317 25348373 25830018 26108605 25315136 21347285 21719571 26334808 29403074 27129103 26823975
12364791 14747013 17210077 24508170 30249741 18362917 19820115 17994087 15632085 26369729 20353571 18057021 28648823 17550304 20075255 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 22936249 26109357 26109356 24495485 20566863 20798317 25348373 25830018 26108605 25315136 21347285 21719571 26334808 29403074 27129103 26823975
EMBL
BABH01006724
AGBW02013811
OWR42745.1
ODYU01009477
SOQ53948.1
NWSH01000159
+ More
PCG78943.1 AK401849 KQ459463 BAM18471.1 KPJ00599.1 KQ461108 KPJ09134.1 GAIX01000803 JAA91757.1 GFDF01011227 JAV02857.1 AAAB01008960 EAA11808.5 APCN01001522 AJWK01007525 AXCN02000352 KK107455 QOIP01000010 EZA50433.1 RLU17667.1 DS497723 EFA12921.1 CH902618 EDV40574.1 OUUW01000002 SPP77522.1 JXJN01009733 CH379070 EAL30186.1 CH479244 EDW37721.1 GGFL01004661 MBW68839.1 CH963847 EDW73398.1 GDAI01002014 JAI15589.1 CH933809 EDW17974.1 CCAG010018738 EZ423180 ADD19456.1 CP012525 ALC43772.1 CH940647 EDW69216.1 KRF84240.1 JR044251 AEY59852.1 KQ414633 KOC67037.1 GGFK01009804 MBW43125.1 GGFM01003129 MBW23880.1 GANO01004841 JAB55030.1 NNAY01002011 OXU22314.1 CM000159 EDW93738.1 AE014296 ACZ94640.2 KQ777311 OAD52138.1 CH954178 EDV50495.1 CH480817 EDW50818.1 CM000363 CM002912 EDX09546.1 KMY98057.1 AY060444 KX531886 AAF50580.1 AAL25483.1 ANY27696.1 CH916366 EDV96142.1 GAMC01001965 GAMC01001964 GAMC01001963 JAC04591.1 DS235854 EEB18864.1 GL445914 EFN88836.1 LBMM01000545 KMQ98093.1 GGFJ01010429 MBW59570.1 GDHF01032705 GDHF01026108 JAI19609.1 JAI26206.1 GAKP01020465 GAKP01020460 GAKP01020457 GAKP01020454 JAC38495.1 KQ434846 KZC08353.1 GAMD01003056 JAA98534.1 KZ288364 PBC26873.1 UFQS01000156 UFQT01000156 SSX00614.1 SSX20994.1 GBXI01009728 GBXI01005628 JAD04564.1 JAD08664.1 LJIG01009236 KRT83484.1 JRES01001180 KNC24728.1 GGFJ01010430 MBW59571.1 GFDL01010114 JAV24931.1 KQ978023 KYM97909.1 KQ976401 KYM92566.1 ADTU01013183 GL435569 EFN73110.1 GL888182 EGI65703.1 GBGD01003423 JAC85466.1 PYGN01000012 PSN57894.1 GDKW01000087 JAI56508.1 DS231949 EDS28529.1 GECL01000804 JAP05320.1 GGFL01004684 MBW68862.1 KQ435729 KOX77620.1 GDHC01020169 GDHC01004527 JAP98459.1 JAQ14102.1 KK854516 PTY16506.1 KQ981490 KYN41038.1
PCG78943.1 AK401849 KQ459463 BAM18471.1 KPJ00599.1 KQ461108 KPJ09134.1 GAIX01000803 JAA91757.1 GFDF01011227 JAV02857.1 AAAB01008960 EAA11808.5 APCN01001522 AJWK01007525 AXCN02000352 KK107455 QOIP01000010 EZA50433.1 RLU17667.1 DS497723 EFA12921.1 CH902618 EDV40574.1 OUUW01000002 SPP77522.1 JXJN01009733 CH379070 EAL30186.1 CH479244 EDW37721.1 GGFL01004661 MBW68839.1 CH963847 EDW73398.1 GDAI01002014 JAI15589.1 CH933809 EDW17974.1 CCAG010018738 EZ423180 ADD19456.1 CP012525 ALC43772.1 CH940647 EDW69216.1 KRF84240.1 JR044251 AEY59852.1 KQ414633 KOC67037.1 GGFK01009804 MBW43125.1 GGFM01003129 MBW23880.1 GANO01004841 JAB55030.1 NNAY01002011 OXU22314.1 CM000159 EDW93738.1 AE014296 ACZ94640.2 KQ777311 OAD52138.1 CH954178 EDV50495.1 CH480817 EDW50818.1 CM000363 CM002912 EDX09546.1 KMY98057.1 AY060444 KX531886 AAF50580.1 AAL25483.1 ANY27696.1 CH916366 EDV96142.1 GAMC01001965 GAMC01001964 GAMC01001963 JAC04591.1 DS235854 EEB18864.1 GL445914 EFN88836.1 LBMM01000545 KMQ98093.1 GGFJ01010429 MBW59570.1 GDHF01032705 GDHF01026108 JAI19609.1 JAI26206.1 GAKP01020465 GAKP01020460 GAKP01020457 GAKP01020454 JAC38495.1 KQ434846 KZC08353.1 GAMD01003056 JAA98534.1 KZ288364 PBC26873.1 UFQS01000156 UFQT01000156 SSX00614.1 SSX20994.1 GBXI01009728 GBXI01005628 JAD04564.1 JAD08664.1 LJIG01009236 KRT83484.1 JRES01001180 KNC24728.1 GGFJ01010430 MBW59571.1 GFDL01010114 JAV24931.1 KQ978023 KYM97909.1 KQ976401 KYM92566.1 ADTU01013183 GL435569 EFN73110.1 GL888182 EGI65703.1 GBGD01003423 JAC85466.1 PYGN01000012 PSN57894.1 GDKW01000087 JAI56508.1 DS231949 EDS28529.1 GECL01000804 JAP05320.1 GGFL01004684 MBW68862.1 KQ435729 KOX77620.1 GDHC01020169 GDHC01004527 JAP98459.1 JAQ14102.1 KK854516 PTY16506.1 KQ981490 KYN41038.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000076408
+ More
UP000075885 UP000007062 UP000075840 UP000092461 UP000075886 UP000053097 UP000279307 UP000007266 UP000007801 UP000268350 UP000092443 UP000092460 UP000001819 UP000008744 UP000007798 UP000009192 UP000078200 UP000092445 UP000092444 UP000092553 UP000005203 UP000008792 UP000069272 UP000053825 UP000215335 UP000002282 UP000002358 UP000000803 UP000075880 UP000008711 UP000001292 UP000000304 UP000001070 UP000009046 UP000008237 UP000036403 UP000076502 UP000242457 UP000037069 UP000095301 UP000078542 UP000078540 UP000005205 UP000000311 UP000007755 UP000245037 UP000095300 UP000002320 UP000053105 UP000078541
UP000075885 UP000007062 UP000075840 UP000092461 UP000075886 UP000053097 UP000279307 UP000007266 UP000007801 UP000268350 UP000092443 UP000092460 UP000001819 UP000008744 UP000007798 UP000009192 UP000078200 UP000092445 UP000092444 UP000092553 UP000005203 UP000008792 UP000069272 UP000053825 UP000215335 UP000002282 UP000002358 UP000000803 UP000075880 UP000008711 UP000001292 UP000000304 UP000001070 UP000009046 UP000008237 UP000036403 UP000076502 UP000242457 UP000037069 UP000095301 UP000078542 UP000078540 UP000005205 UP000000311 UP000007755 UP000245037 UP000095300 UP000002320 UP000053105 UP000078541
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9ITN6
A0A212EMM5
A0A2H1WLP4
A0A2A4K594
I4DKN1
A0A194QUZ0
+ More
S4PZX0 A0A182Y1C1 A0A3F2YWJ7 A0A1L8D932 Q7PNP9 A0A2C9GPQ3 A0A1B0CD90 A0A182Q6I8 A0A026W334 D7EJZ1 B3M569 A0A3B0JBV0 A0A1A9YEH2 A0A1B0B7X0 Q29E76 B4HAT9 A0A2M4CU88 B4MLE0 A0A0K8TMM8 B4KZW2 A0A1A9V479 A0A1A9ZS28 D3TP28 A0A0M4EKI5 A0A088A5C5 B4LFB8 A0A182FG97 V9IGQ3 A0A0L7R827 A0A2M4AQQ3 A0A2M3Z5U4 U5ELL6 A0A232EVD5 B4PK58 K7J651 E1JIE5 A0A182J000 A0A310SET5 B3NFF3 B4HV47 B4QKC3 Q9VS49 B4J371 W8BTA3 E0VZQ8 E2B647 A0A0J7L6C2 A0A2M4C2M7 A0A0K8TYQ0 A0A034V9N7 A0A154P932 T1DFS2 A0A2A3E537 A0A336LSN1 A0A0A1XB99 A0A0T6B7Z6 A0A0L0BXD7 A0A2M4C336 A0A1I8MM87 A0A1Q3FBI2 A0A195CAL2 A0A195BX83 A0A158NEA1 E2A0A7 F4WJC5 A0A069DP63 A0A2P8ZN15 A0A0P4W0U6 A0A1I8Q4L9 B0WIK6 A0A0V0GC25 A0A2M4CU42 A0A0M9A5G3 A0A146M515 A0A2R7WA26 A0A195FKU4
S4PZX0 A0A182Y1C1 A0A3F2YWJ7 A0A1L8D932 Q7PNP9 A0A2C9GPQ3 A0A1B0CD90 A0A182Q6I8 A0A026W334 D7EJZ1 B3M569 A0A3B0JBV0 A0A1A9YEH2 A0A1B0B7X0 Q29E76 B4HAT9 A0A2M4CU88 B4MLE0 A0A0K8TMM8 B4KZW2 A0A1A9V479 A0A1A9ZS28 D3TP28 A0A0M4EKI5 A0A088A5C5 B4LFB8 A0A182FG97 V9IGQ3 A0A0L7R827 A0A2M4AQQ3 A0A2M3Z5U4 U5ELL6 A0A232EVD5 B4PK58 K7J651 E1JIE5 A0A182J000 A0A310SET5 B3NFF3 B4HV47 B4QKC3 Q9VS49 B4J371 W8BTA3 E0VZQ8 E2B647 A0A0J7L6C2 A0A2M4C2M7 A0A0K8TYQ0 A0A034V9N7 A0A154P932 T1DFS2 A0A2A3E537 A0A336LSN1 A0A0A1XB99 A0A0T6B7Z6 A0A0L0BXD7 A0A2M4C336 A0A1I8MM87 A0A1Q3FBI2 A0A195CAL2 A0A195BX83 A0A158NEA1 E2A0A7 F4WJC5 A0A069DP63 A0A2P8ZN15 A0A0P4W0U6 A0A1I8Q4L9 B0WIK6 A0A0V0GC25 A0A2M4CU42 A0A0M9A5G3 A0A146M515 A0A2R7WA26 A0A195FKU4
Ontologies
PANTHER
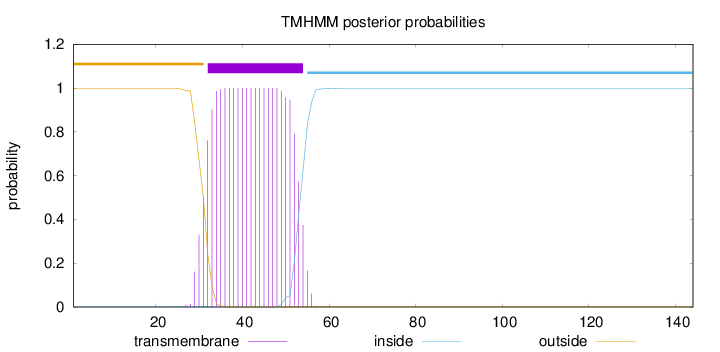
Topology
Length:
144
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.52727
Exp number, first 60 AAs:
22.52673
Total prob of N-in:
0.00139
POSSIBLE N-term signal
sequence
outside
1 - 31
TMhelix
32 - 54
inside
55 - 144
Population Genetic Test Statistics
Pi
0.71691
Theta
2.989391
Tajima's D
-1.980972
CLR
8.351494
CSRT
0.00939953002349883
Interpretation
Possibly Positive selection
