Gene
KWMTBOMO00170
Pre Gene Modal
BGIBMGA000610
Annotation
PREDICTED:_uncharacterized_protein_LOC106133963_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.499 PlasmaMembrane Reliability : 1.231
Sequence
CDS
ATGAAACCACTATCGCTGGATGTTATTTCAAGATACCTTACTAAAAATGTTATAGCGGAAGTTTTTAAATTGAAGACAGACAGCGATGAAGACATAGAAAATATTGAATTAACAAAAGCAGCACCAAATGGCGAAGGTCTCCTCAGTGCAGTCTACAGGATCAAAGTTACAGGCAAGAGATATTCCACATCCTTTATCGGGAAGGGTTTCGTTGACAATGAACGCATCAAAAACTCTTTAAACTGTGCCGAACTATTCAGAAGAGAGGCATTGTTTTACTCTGATGTGCTACCTATACTAGTCGATTTGCAGCAATCGTTAGGTGCGAGCGAATGCATTCAGAACAGCGTGCCTATTTGTTATGGTTGTTATCTGGACGGCGTTAATGATTATATCGTTCTAGAAGATCTTGGCGAAAGTCGCTGTAAATCTTTAACGAAACACCCTACAAAGTGCGAACGGGATTCAGTTCTAGAGACACTAGCACATTTGCACGCAGTATCGATGACTTTGCGAATCAAGAAGCCCGATGAATTCAAAAGGATTGCCGATTTAATTCCAGAAGCGTACTATACTGAAAAAAATCGTGATTGGTATTCGAAATTTCAACACAAAGCTGTAATGATAGATCAAGCGATTTTAATTGAACACGAAGATCCATCGTCTGTTTACTATAAAAAATATAACGAGCTGCTCGGTCAAGACGTGTACGGGTGCTTGATCGAAATGACGACGACTCGTGGAGATTTCACTGTGATCAATCACGGAGACTCTTGGACTCCGAATTTTCTTCGTTCTAAAGGCAGAACTGTTGCGATTGATTTTCAGATGATACGATGCGCTTCGCCAGTTACGGATCTAATGCTTTTCTTGTTTATGTGCACAAACGTTTGCTCAAATGTCGAACAGTTTGAGGAAGCTCTCGACCATTATTATAATTTTTTAGATTATTTTTTGAAAGACATGGGCATGGACGTCGCAATTGTCTTCCCAAGAGACGCTTTCGAAAACGAACTCAAAATATACGGAAAGTTCGGATTTCTTACATGCCTGACGTCAATTCCGCTCTTAGCAAACGAGGGGACCAGTGCGTTACAATTGACAGAAGATTTTCCAGAACATAGTCGAATACCATTAGAAAAACTATGGGACATTTCACCTATCAAAAACGAGCAGTATCAGATGAGATTTGTGAACGCTTTAAGACTAGCTGTAGATGCGAATCTTATTTGA
Protein
MKPLSLDVISRYLTKNVIAEVFKLKTDSDEDIENIELTKAAPNGEGLLSAVYRIKVTGKRYSTSFIGKGFVDNERIKNSLNCAELFRREALFYSDVLPILVDLQQSLGASECIQNSVPICYGCYLDGVNDYIVLEDLGESRCKSLTKHPTKCERDSVLETLAHLHAVSMTLRIKKPDEFKRIADLIPEAYYTEKNRDWYSKFQHKAVMIDQAILIEHEDPSSVYYKKYNELLGQDVYGCLIEMTTTRGDFTVINHGDSWTPNFLRSKGRTVAIDFQMIRCASPVTDLMLFLFMCTNVCSNVEQFEEALDHYYNFLDYFLKDMGMDVAIVFPRDAFENELKIYGKFGFLTCLTSIPLLANEGTSALQLTEDFPEHSRIPLEKLWDISPIKNEQYQMRFVNALRLAVDANLI
Summary
Similarity
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Kinesin family.
Uniprot
H9ITN0
A0A2W1BU18
A0A2A4JM67
A0A2H1VP32
A0A194QV60
A0A0L7LF36
+ More
A0A194PS91 A0A182N5A7 F5HL17 A0A2J7QMA2 A0A3F2Z0C6 A0A182HJA1 A0A182XE21 Q7QBD5 A0A3F2Z0C7 A0A182S5M1 A0A1I8JUR4 A0A182Q9T9 A0A182PE46 A0A336LPX7 A0A182M746 A0A0C9R214 A0A182J0Y1 A0A194Q5F4 A0A182K9D3 A0A182LCI5 A0A182TV21 H9JWQ7 A0A232EF14 A0A1B0CGM7 U5ET49 A0A0L0CKN4 A0A194QVA2 A0A2P8ZFB9 A0A182VRI2 K7IS29 A0A0M4EUW7 A0A2M4BQ86 A0A212EPF3 A0A084WSL8 A0A2H1X1G0 Q29GS3 A0A154NZB0 A0A2M3ZIR7 D1ZZT4 W5J6B9 B4GUV1 B3MZG8 A0A1I8NQR5 A0A2M4BPD3 B4L3L2 B4MD83 B4IK01 A0A1A9V5G1 B4R6L4 A0A3B0KJ84 B3NUL1 A0A2W1BGJ3 A0A1B0FD05 A0A194Q0V2 A0A1L8DN26 A0A1L8DN21 A0A088A0R6 A0A034WGQ0 A0A336M4I1 A0A1W4USA3 A0A1Y1MPW9 Q9W3C3 T1PB05 A0A195F6P1 D1ZZT3 A0A0K8W2E8 B4Q013 A0A158NNK3 A0A182FSK7 B4JJ52 A0A0K8VKA0 A0A0A1XEX0 A0A195B386 F4WQK7 Q17DW7 W8BSE4 A0A1A9XAV3 A0A182GTK0 A0A0K8WCV9 A0A023ERZ5 A0A2A3EQR7 A0A151X957 A0A0J7MYY7 B4MJ43 A0A195DKP4 A0A232EF01 A0A232EHX7 A0A1Y1MPA5 A0A2A4JMI3 A0A0N0BIA1 A0A3L8E100
A0A194PS91 A0A182N5A7 F5HL17 A0A2J7QMA2 A0A3F2Z0C6 A0A182HJA1 A0A182XE21 Q7QBD5 A0A3F2Z0C7 A0A182S5M1 A0A1I8JUR4 A0A182Q9T9 A0A182PE46 A0A336LPX7 A0A182M746 A0A0C9R214 A0A182J0Y1 A0A194Q5F4 A0A182K9D3 A0A182LCI5 A0A182TV21 H9JWQ7 A0A232EF14 A0A1B0CGM7 U5ET49 A0A0L0CKN4 A0A194QVA2 A0A2P8ZFB9 A0A182VRI2 K7IS29 A0A0M4EUW7 A0A2M4BQ86 A0A212EPF3 A0A084WSL8 A0A2H1X1G0 Q29GS3 A0A154NZB0 A0A2M3ZIR7 D1ZZT4 W5J6B9 B4GUV1 B3MZG8 A0A1I8NQR5 A0A2M4BPD3 B4L3L2 B4MD83 B4IK01 A0A1A9V5G1 B4R6L4 A0A3B0KJ84 B3NUL1 A0A2W1BGJ3 A0A1B0FD05 A0A194Q0V2 A0A1L8DN26 A0A1L8DN21 A0A088A0R6 A0A034WGQ0 A0A336M4I1 A0A1W4USA3 A0A1Y1MPW9 Q9W3C3 T1PB05 A0A195F6P1 D1ZZT3 A0A0K8W2E8 B4Q013 A0A158NNK3 A0A182FSK7 B4JJ52 A0A0K8VKA0 A0A0A1XEX0 A0A195B386 F4WQK7 Q17DW7 W8BSE4 A0A1A9XAV3 A0A182GTK0 A0A0K8WCV9 A0A023ERZ5 A0A2A3EQR7 A0A151X957 A0A0J7MYY7 B4MJ43 A0A195DKP4 A0A232EF01 A0A232EHX7 A0A1Y1MPA5 A0A2A4JMI3 A0A0N0BIA1 A0A3L8E100
Pubmed
19121390
28756777
26354079
26227816
12364791
14747013
+ More
17210077 20966253 28648823 26108605 29403074 20075255 22118469 24438588 15632085 17994087 23185243 18362917 19820115 20920257 23761445 18057021 25348373 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 21347285 25830018 21719571 17510324 24495485 26483478 24945155 30249741
17210077 20966253 28648823 26108605 29403074 20075255 22118469 24438588 15632085 17994087 23185243 18362917 19820115 20920257 23761445 18057021 25348373 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 21347285 25830018 21719571 17510324 24495485 26483478 24945155 30249741
EMBL
BABH01006695
KZ149901
PZC78509.1
NWSH01001050
PCG72919.1
ODYU01003434
+ More
SOQ42202.1 KQ461108 KPJ09204.1 JTDY01001461 KOB73811.1 KQ459594 KPI96182.1 AAAB01008879 EGK96978.1 NEVH01013215 PNF29697.1 APCN01002134 EAA08488.3 AXCN02000148 UFQT01000109 SSX20136.1 AXCM01004215 GBYB01000901 JAG70668.1 KQ459583 KPI98625.1 BABH01018468 NNAY01005152 OXU16941.1 AJWK01011468 GANO01004535 JAB55336.1 JRES01000348 KNC32029.1 KQ461103 KPJ09463.1 PYGN01000075 PSN55206.1 CP012528 ALC49638.1 GGFJ01005787 MBW54928.1 AGBW02013479 OWR43359.1 ATLV01026615 KE525415 KFB53212.1 ODYU01012709 SOQ59151.1 CH379064 EAL32036.1 KRT06790.1 KQ434783 KZC04967.1 GGFM01007670 MBW28421.1 KQ971338 EFA02417.1 ADMH02002074 ETN59546.1 CH479192 EDW26488.1 CH902635 EDV33769.1 GGFJ01005788 MBW54929.1 CH933810 EDW07140.2 CH940660 EDW58155.1 CH480850 EDW51357.1 CM000366 EDX17444.1 OUUW01000015 SPP88610.1 CH954180 EDV46334.1 KQS29935.1 KZ150238 PZC71946.1 CCAG010013058 KPI98624.1 GFDF01006232 JAV07852.1 GFDF01006231 JAV07853.1 GAKP01005078 JAC53874.1 UFQS01000550 UFQT01000550 SSX04802.1 SSX25165.1 GEZM01028014 GEZM01028013 JAV86515.1 AE014298 AY118900 AAF46408.1 AAM50760.1 KA645128 AFP59757.1 KQ981744 KYN36265.1 EFA02416.1 GDHF01006961 JAI45353.1 CM000162 EDX02219.1 KRK06644.1 ADTU01021631 CH916370 EDV99604.1 GDHF01012998 JAI39316.1 GBXI01004358 JAD09934.1 KQ976641 KYM78730.1 GL888273 EGI63534.1 CH477290 EAT44634.1 GAMC01010354 GAMC01010353 GAMC01010350 JAB96201.1 JXUM01087313 KQ563625 KXJ73581.1 GDHF01003392 JAI48922.1 GAPW01001613 JAC11985.1 KZ288201 PBC33566.1 KQ982383 KYQ56907.1 LBMM01013392 KMQ85660.1 CH963738 EDW72132.1 KQ980765 KYN13448.1 OXU16940.1 NNAY01004383 OXU17970.1 GEZM01028018 GEZM01028015 JAV86510.1 NWSH01001035 PCG72996.1 KQ435732 KOX77392.1 QOIP01000001 RLU26400.1
SOQ42202.1 KQ461108 KPJ09204.1 JTDY01001461 KOB73811.1 KQ459594 KPI96182.1 AAAB01008879 EGK96978.1 NEVH01013215 PNF29697.1 APCN01002134 EAA08488.3 AXCN02000148 UFQT01000109 SSX20136.1 AXCM01004215 GBYB01000901 JAG70668.1 KQ459583 KPI98625.1 BABH01018468 NNAY01005152 OXU16941.1 AJWK01011468 GANO01004535 JAB55336.1 JRES01000348 KNC32029.1 KQ461103 KPJ09463.1 PYGN01000075 PSN55206.1 CP012528 ALC49638.1 GGFJ01005787 MBW54928.1 AGBW02013479 OWR43359.1 ATLV01026615 KE525415 KFB53212.1 ODYU01012709 SOQ59151.1 CH379064 EAL32036.1 KRT06790.1 KQ434783 KZC04967.1 GGFM01007670 MBW28421.1 KQ971338 EFA02417.1 ADMH02002074 ETN59546.1 CH479192 EDW26488.1 CH902635 EDV33769.1 GGFJ01005788 MBW54929.1 CH933810 EDW07140.2 CH940660 EDW58155.1 CH480850 EDW51357.1 CM000366 EDX17444.1 OUUW01000015 SPP88610.1 CH954180 EDV46334.1 KQS29935.1 KZ150238 PZC71946.1 CCAG010013058 KPI98624.1 GFDF01006232 JAV07852.1 GFDF01006231 JAV07853.1 GAKP01005078 JAC53874.1 UFQS01000550 UFQT01000550 SSX04802.1 SSX25165.1 GEZM01028014 GEZM01028013 JAV86515.1 AE014298 AY118900 AAF46408.1 AAM50760.1 KA645128 AFP59757.1 KQ981744 KYN36265.1 EFA02416.1 GDHF01006961 JAI45353.1 CM000162 EDX02219.1 KRK06644.1 ADTU01021631 CH916370 EDV99604.1 GDHF01012998 JAI39316.1 GBXI01004358 JAD09934.1 KQ976641 KYM78730.1 GL888273 EGI63534.1 CH477290 EAT44634.1 GAMC01010354 GAMC01010353 GAMC01010350 JAB96201.1 JXUM01087313 KQ563625 KXJ73581.1 GDHF01003392 JAI48922.1 GAPW01001613 JAC11985.1 KZ288201 PBC33566.1 KQ982383 KYQ56907.1 LBMM01013392 KMQ85660.1 CH963738 EDW72132.1 KQ980765 KYN13448.1 OXU16940.1 NNAY01004383 OXU17970.1 GEZM01028018 GEZM01028015 JAV86510.1 NWSH01001035 PCG72996.1 KQ435732 KOX77392.1 QOIP01000001 RLU26400.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000053268
UP000075884
+ More
UP000007062 UP000235965 UP000075903 UP000075840 UP000076407 UP000075901 UP000075900 UP000075886 UP000075885 UP000075883 UP000075880 UP000075881 UP000075882 UP000075902 UP000215335 UP000092461 UP000037069 UP000245037 UP000075920 UP000002358 UP000092553 UP000007151 UP000030765 UP000001819 UP000076502 UP000007266 UP000000673 UP000008744 UP000007801 UP000095300 UP000009192 UP000008792 UP000001292 UP000078200 UP000000304 UP000268350 UP000008711 UP000092444 UP000005203 UP000192221 UP000000803 UP000095301 UP000078541 UP000002282 UP000005205 UP000069272 UP000001070 UP000078540 UP000007755 UP000008820 UP000092443 UP000069940 UP000249989 UP000242457 UP000075809 UP000036403 UP000007798 UP000078492 UP000053105 UP000279307
UP000007062 UP000235965 UP000075903 UP000075840 UP000076407 UP000075901 UP000075900 UP000075886 UP000075885 UP000075883 UP000075880 UP000075881 UP000075882 UP000075902 UP000215335 UP000092461 UP000037069 UP000245037 UP000075920 UP000002358 UP000092553 UP000007151 UP000030765 UP000001819 UP000076502 UP000007266 UP000000673 UP000008744 UP000007801 UP000095300 UP000009192 UP000008792 UP000001292 UP000078200 UP000000304 UP000268350 UP000008711 UP000092444 UP000005203 UP000192221 UP000000803 UP000095301 UP000078541 UP000002282 UP000005205 UP000069272 UP000001070 UP000078540 UP000007755 UP000008820 UP000092443 UP000069940 UP000249989 UP000242457 UP000075809 UP000036403 UP000007798 UP000078492 UP000053105 UP000279307
Interpro
IPR015897
CHK_kinase-like
+ More
IPR011009 Kinase-like_dom_sf
IPR004119 EcKinase-like
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR039647 EF_hand_pair_protein_CML-like
IPR018247 EF_Hand_1_Ca_BS
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR027417 P-loop_NTPase
IPR038105 Kif23_Arf-bd_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR011009 Kinase-like_dom_sf
IPR004119 EcKinase-like
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR039647 EF_hand_pair_protein_CML-like
IPR018247 EF_Hand_1_Ca_BS
IPR001752 Kinesin_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR027417 P-loop_NTPase
IPR038105 Kif23_Arf-bd_sf
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
Gene 3D
ProteinModelPortal
H9ITN0
A0A2W1BU18
A0A2A4JM67
A0A2H1VP32
A0A194QV60
A0A0L7LF36
+ More
A0A194PS91 A0A182N5A7 F5HL17 A0A2J7QMA2 A0A3F2Z0C6 A0A182HJA1 A0A182XE21 Q7QBD5 A0A3F2Z0C7 A0A182S5M1 A0A1I8JUR4 A0A182Q9T9 A0A182PE46 A0A336LPX7 A0A182M746 A0A0C9R214 A0A182J0Y1 A0A194Q5F4 A0A182K9D3 A0A182LCI5 A0A182TV21 H9JWQ7 A0A232EF14 A0A1B0CGM7 U5ET49 A0A0L0CKN4 A0A194QVA2 A0A2P8ZFB9 A0A182VRI2 K7IS29 A0A0M4EUW7 A0A2M4BQ86 A0A212EPF3 A0A084WSL8 A0A2H1X1G0 Q29GS3 A0A154NZB0 A0A2M3ZIR7 D1ZZT4 W5J6B9 B4GUV1 B3MZG8 A0A1I8NQR5 A0A2M4BPD3 B4L3L2 B4MD83 B4IK01 A0A1A9V5G1 B4R6L4 A0A3B0KJ84 B3NUL1 A0A2W1BGJ3 A0A1B0FD05 A0A194Q0V2 A0A1L8DN26 A0A1L8DN21 A0A088A0R6 A0A034WGQ0 A0A336M4I1 A0A1W4USA3 A0A1Y1MPW9 Q9W3C3 T1PB05 A0A195F6P1 D1ZZT3 A0A0K8W2E8 B4Q013 A0A158NNK3 A0A182FSK7 B4JJ52 A0A0K8VKA0 A0A0A1XEX0 A0A195B386 F4WQK7 Q17DW7 W8BSE4 A0A1A9XAV3 A0A182GTK0 A0A0K8WCV9 A0A023ERZ5 A0A2A3EQR7 A0A151X957 A0A0J7MYY7 B4MJ43 A0A195DKP4 A0A232EF01 A0A232EHX7 A0A1Y1MPA5 A0A2A4JMI3 A0A0N0BIA1 A0A3L8E100
A0A194PS91 A0A182N5A7 F5HL17 A0A2J7QMA2 A0A3F2Z0C6 A0A182HJA1 A0A182XE21 Q7QBD5 A0A3F2Z0C7 A0A182S5M1 A0A1I8JUR4 A0A182Q9T9 A0A182PE46 A0A336LPX7 A0A182M746 A0A0C9R214 A0A182J0Y1 A0A194Q5F4 A0A182K9D3 A0A182LCI5 A0A182TV21 H9JWQ7 A0A232EF14 A0A1B0CGM7 U5ET49 A0A0L0CKN4 A0A194QVA2 A0A2P8ZFB9 A0A182VRI2 K7IS29 A0A0M4EUW7 A0A2M4BQ86 A0A212EPF3 A0A084WSL8 A0A2H1X1G0 Q29GS3 A0A154NZB0 A0A2M3ZIR7 D1ZZT4 W5J6B9 B4GUV1 B3MZG8 A0A1I8NQR5 A0A2M4BPD3 B4L3L2 B4MD83 B4IK01 A0A1A9V5G1 B4R6L4 A0A3B0KJ84 B3NUL1 A0A2W1BGJ3 A0A1B0FD05 A0A194Q0V2 A0A1L8DN26 A0A1L8DN21 A0A088A0R6 A0A034WGQ0 A0A336M4I1 A0A1W4USA3 A0A1Y1MPW9 Q9W3C3 T1PB05 A0A195F6P1 D1ZZT3 A0A0K8W2E8 B4Q013 A0A158NNK3 A0A182FSK7 B4JJ52 A0A0K8VKA0 A0A0A1XEX0 A0A195B386 F4WQK7 Q17DW7 W8BSE4 A0A1A9XAV3 A0A182GTK0 A0A0K8WCV9 A0A023ERZ5 A0A2A3EQR7 A0A151X957 A0A0J7MYY7 B4MJ43 A0A195DKP4 A0A232EF01 A0A232EHX7 A0A1Y1MPA5 A0A2A4JMI3 A0A0N0BIA1 A0A3L8E100
Ontologies
GO
PANTHER
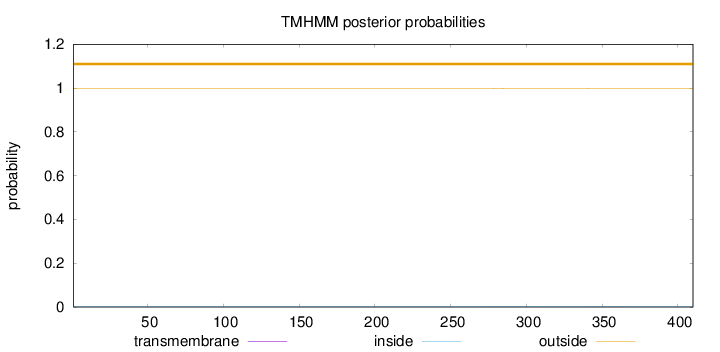
Topology
Length:
410
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02744
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00190
outside
1 - 410
Population Genetic Test Statistics
Pi
14.867201
Theta
13.589377
Tajima's D
0.294245
CLR
0.0035
CSRT
0.463276836158192
Interpretation
Uncertain
