Gene
KWMTBOMO00168
Pre Gene Modal
BGIBMGA000608
Annotation
uncharacterized_protein_LOC101741444_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.655
Sequence
CDS
ATGATACGGAACAAGAAGCCACCATTCTCTATGAATCGTTTTACGGGAATGCTGATGGTTGGTTATTCCTGGGGTCAAATGATGATGGTAGCAGTGCCATCTGATGAAATATGGGGTATCAATTTTAGATATCTCAACTACCTCATTCCTTTGGTTGCTGCACTAGGTGTCTGGACAGTAGGCAACATTGGTCACGAGCAAGGAACCTTGAAGTGGCCTCTCTTAGCTGCATATGTGTCCTATCCTCTTCGCTACTTTATATATGATGAAACAGTATTTTTCACTATCATGGTGTTGTCTGCAGCATTAGCATTTGACTCCTTCTCAAAGCAGTGGCGCAGAACTCCAAGAAGGCATCGTTTTGTTAAGAGAGTGATTGTCCTCGGTGTGTGTGCTTGCCTTTACTTATCCCTATGGTGTAGCTACTTGTACTTCAATGGCACGATCACTGACAGTGAAGGCGACGAGGTTCCAATCTACGAAGCATTGCATCACTTCTTTACAAGTCCTTGGTGGCTTGATGTGAAGCAGTGCCTTATTGATACATACCAGTATGCACAGCATCATGGCTGGTATGAAGTGTGGAAACAAATTATTGATCTGTCTGATCCTCATGGTGAACAAAACGCATACAAGGTGCTCGGGCTGGGCCCTGAAGCTACACAACAAGAGATAACTTCTACTTGGCGAAGATTGTCCAGAGAGCATCATCCCGACAAAGTCAAGGATGAGAACGGACGCCGCGCTGCCCAAGAACGATTCATGGAAATACAACAAGCTTACGAGATCTTGTCCAATAGCAAGCACAGAAGAAATCGACGCAATAAGAAAGATAACTCGGAGTAA
Protein
MIRNKKPPFSMNRFTGMLMVGYSWGQMMMVAVPSDEIWGINFRYLNYLIPLVAALGVWTVGNIGHEQGTLKWPLLAAYVSYPLRYFIYDETVFFTIMVLSAALAFDSFSKQWRRTPRRHRFVKRVIVLGVCACLYLSLWCSYLYFNGTITDSEGDEVPIYEALHHFFTSPWWLDVKQCLIDTYQYAQHHGWYEVWKQIIDLSDPHGEQNAYKVLGLGPEATQQEITSTWRRLSREHHPDKVKDENGRRAAQERFMEIQQAYEILSNSKHRRNRRNKKDNSE
Summary
Uniprot
H6VTQ8
A0A194PYF7
A0A194R027
A0A2A4JJX6
A0A212FM98
A0A2H1VN18
+ More
D6WIK7 A0A1W4WE44 A0A1Y1MMX0 A0A1J1IQG1 N6T5Z5 A0A067RPR7 A0A182GIN4 E0VZD7 A0A182GPW6 A0A2P8Z3X9 A0A336LMD3 A0A336M1U2 U4UG80 Q1HRR7 Q16UA2 A0A182WLK1 A0A182YKS5 A0A182RDM9 U5EZU4 A0A182J498 A0A182QAH7 A0A2M4CW11 A0A182ST93 A0A2M4BPM7 A0A2M3ZF21 A0A2M3YXG4 A0A2M4CW10 A0A1L8DTL0 A0A2M4A064 A0A2M3YXG5 A0A084WEL7 A0A2M3ZZ44 B0XGC5 A0A182TGZ2 A0A182I2K5 A0A182PCT1 A0A182V5G4 A0A182NK41 A0A182KQG8 A0A1Q3FDP8 Q7PTB4 A0A1S3D130 A0A1I8MEP3 A0A182LVJ1 A0A1I8Q1R3 A0A1B0G7E9 A0A182JP58 A0A0L0C1R2 F4WG03 A0A1A9XZK4 A0A1B0BYV7 A0A1A9VNL4 A0A195EGD5 A0A1A9ZUZ9 B4NPR9 A0A195BG49 W8CE63 A0A088A3K8 A0A2A3ESE4 A0A151XBM1 A0A0A1WPF6 A0A0K8UCV7 A0A182WX10 A0A1A9W119 A0A2H8TEW0 A0A026WSA5 A0A154P0K0 A0A069DSM4 A0A034VVG9 A0A195F0L6 A0A0V0G8R2 A0A0P4VWA9 A0A195BYS4 A0A182FH63 A0A2S2PCD9 A0A0L7QS91 Q9VX95 A0A2S2QN34 E9IN27 J9K8L7 A0A1W4VVB8 B3NWA2 T1I789 E2BVB1 A0A0M4FA27 B3N0B7 B4L230 E2ASK8 B4Q1X4 B4M1K0 B4JN54 A0A232EUN0 K7IN06 A0A1B0EZR6
D6WIK7 A0A1W4WE44 A0A1Y1MMX0 A0A1J1IQG1 N6T5Z5 A0A067RPR7 A0A182GIN4 E0VZD7 A0A182GPW6 A0A2P8Z3X9 A0A336LMD3 A0A336M1U2 U4UG80 Q1HRR7 Q16UA2 A0A182WLK1 A0A182YKS5 A0A182RDM9 U5EZU4 A0A182J498 A0A182QAH7 A0A2M4CW11 A0A182ST93 A0A2M4BPM7 A0A2M3ZF21 A0A2M3YXG4 A0A2M4CW10 A0A1L8DTL0 A0A2M4A064 A0A2M3YXG5 A0A084WEL7 A0A2M3ZZ44 B0XGC5 A0A182TGZ2 A0A182I2K5 A0A182PCT1 A0A182V5G4 A0A182NK41 A0A182KQG8 A0A1Q3FDP8 Q7PTB4 A0A1S3D130 A0A1I8MEP3 A0A182LVJ1 A0A1I8Q1R3 A0A1B0G7E9 A0A182JP58 A0A0L0C1R2 F4WG03 A0A1A9XZK4 A0A1B0BYV7 A0A1A9VNL4 A0A195EGD5 A0A1A9ZUZ9 B4NPR9 A0A195BG49 W8CE63 A0A088A3K8 A0A2A3ESE4 A0A151XBM1 A0A0A1WPF6 A0A0K8UCV7 A0A182WX10 A0A1A9W119 A0A2H8TEW0 A0A026WSA5 A0A154P0K0 A0A069DSM4 A0A034VVG9 A0A195F0L6 A0A0V0G8R2 A0A0P4VWA9 A0A195BYS4 A0A182FH63 A0A2S2PCD9 A0A0L7QS91 Q9VX95 A0A2S2QN34 E9IN27 J9K8L7 A0A1W4VVB8 B3NWA2 T1I789 E2BVB1 A0A0M4FA27 B3N0B7 B4L230 E2ASK8 B4Q1X4 B4M1K0 B4JN54 A0A232EUN0 K7IN06 A0A1B0EZR6
Pubmed
19121390
26434795
26354079
22118469
18362917
19820115
+ More
28004739 23537049 24845553 26483478 20566863 29403074 17204158 17510324 25244985 24438588 20966253 12364791 14747013 17210077 25315136 26108605 21719571 17994087 24495485 25830018 24508170 30249741 26334808 25348373 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 20798317 18057021 17550304 28648823 20075255
28004739 23537049 24845553 26483478 20566863 29403074 17204158 17510324 25244985 24438588 20966253 12364791 14747013 17210077 25315136 26108605 21719571 17994087 24495485 25830018 24508170 30249741 26334808 25348373 27129103 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21282665 20798317 18057021 17550304 28648823 20075255
EMBL
BABH01006694
JN872893
AFC01232.1
KQ459594
KPI96185.1
KQ461108
+ More
KPJ09201.1 NWSH01001220 PCG72076.1 AGBW02007654 OWR54878.1 ODYU01003434 SOQ42198.1 KQ971321 EFA00002.1 GEZM01026388 JAV87021.1 CVRI01000057 CRL01978.1 APGK01042608 KB741007 ENN75609.1 KK852542 KDR21704.1 JXUM01066602 KQ562407 KXJ75980.1 DS235851 EEB18743.1 JXUM01080377 KQ563182 KXJ74320.1 PYGN01000208 PSN51196.1 UFQT01000075 SSX19404.1 UFQT01000299 SSX22969.1 KB632182 ERL89616.1 DQ440027 CH477755 ABF18060.1 EAT36586.1 CH477627 EAT38114.1 GANO01000130 JAB59741.1 AXCN02000433 GGFL01005338 MBW69516.1 GGFJ01005895 MBW55036.1 GGFM01006403 MBW27154.1 GGFM01000203 MBW20954.1 GGFL01005339 MBW69517.1 GFDF01004291 JAV09793.1 GGFK01000789 MBW34110.1 GGFM01000199 MBW20950.1 ATLV01023228 KE525341 KFB48661.1 GGFK01000506 MBW33827.1 DS233012 EDS27507.1 APCN01000169 GFDL01009371 JAV25674.1 AAAB01008807 EAA04160.5 AXCM01004012 CCAG010021563 JRES01001003 KNC26211.1 GL888128 EGI66770.1 JXJN01022848 KQ978957 KYN26954.1 CH964291 EDW86509.1 KQ976488 KYM83567.1 GAMC01000746 JAC05810.1 KZ288187 PBC34713.1 KQ982320 KYQ57776.1 GBXI01014009 GBXI01008753 JAD00283.1 JAD05539.1 GDHF01028918 GDHF01027850 JAI23396.1 JAI24464.1 GFXV01000814 MBW12619.1 KK107111 QOIP01000011 EZA58930.1 RLU16944.1 KQ434783 KZC04778.1 GBGD01001974 JAC86915.1 GAKP01012855 JAC46097.1 KQ981866 KYN34125.1 GECL01001968 JAP04156.1 GDKW01002703 JAI53892.1 KQ978501 KYM93465.1 GGMR01014490 MBY27109.1 KQ414758 KOC61507.1 AE014298 AY058516 AAF48683.1 AAL13745.1 AHN59825.1 GGMS01009898 MBY79101.1 GL764272 EFZ18014.1 ABLF02024672 CH954180 EDV46441.1 ACPB03016783 GL450824 EFN80403.1 CP012528 ALC49195.1 CH902640 EDV38321.1 CH933810 EDW06770.1 KRF93810.1 KRF93811.1 GL442330 EFN63568.1 CM000162 EDX02549.1 CH940651 EDW65554.1 CH916371 EDV92147.1 NNAY01002105 OXU22070.1 AJVK01034775
KPJ09201.1 NWSH01001220 PCG72076.1 AGBW02007654 OWR54878.1 ODYU01003434 SOQ42198.1 KQ971321 EFA00002.1 GEZM01026388 JAV87021.1 CVRI01000057 CRL01978.1 APGK01042608 KB741007 ENN75609.1 KK852542 KDR21704.1 JXUM01066602 KQ562407 KXJ75980.1 DS235851 EEB18743.1 JXUM01080377 KQ563182 KXJ74320.1 PYGN01000208 PSN51196.1 UFQT01000075 SSX19404.1 UFQT01000299 SSX22969.1 KB632182 ERL89616.1 DQ440027 CH477755 ABF18060.1 EAT36586.1 CH477627 EAT38114.1 GANO01000130 JAB59741.1 AXCN02000433 GGFL01005338 MBW69516.1 GGFJ01005895 MBW55036.1 GGFM01006403 MBW27154.1 GGFM01000203 MBW20954.1 GGFL01005339 MBW69517.1 GFDF01004291 JAV09793.1 GGFK01000789 MBW34110.1 GGFM01000199 MBW20950.1 ATLV01023228 KE525341 KFB48661.1 GGFK01000506 MBW33827.1 DS233012 EDS27507.1 APCN01000169 GFDL01009371 JAV25674.1 AAAB01008807 EAA04160.5 AXCM01004012 CCAG010021563 JRES01001003 KNC26211.1 GL888128 EGI66770.1 JXJN01022848 KQ978957 KYN26954.1 CH964291 EDW86509.1 KQ976488 KYM83567.1 GAMC01000746 JAC05810.1 KZ288187 PBC34713.1 KQ982320 KYQ57776.1 GBXI01014009 GBXI01008753 JAD00283.1 JAD05539.1 GDHF01028918 GDHF01027850 JAI23396.1 JAI24464.1 GFXV01000814 MBW12619.1 KK107111 QOIP01000011 EZA58930.1 RLU16944.1 KQ434783 KZC04778.1 GBGD01001974 JAC86915.1 GAKP01012855 JAC46097.1 KQ981866 KYN34125.1 GECL01001968 JAP04156.1 GDKW01002703 JAI53892.1 KQ978501 KYM93465.1 GGMR01014490 MBY27109.1 KQ414758 KOC61507.1 AE014298 AY058516 AAF48683.1 AAL13745.1 AHN59825.1 GGMS01009898 MBY79101.1 GL764272 EFZ18014.1 ABLF02024672 CH954180 EDV46441.1 ACPB03016783 GL450824 EFN80403.1 CP012528 ALC49195.1 CH902640 EDV38321.1 CH933810 EDW06770.1 KRF93810.1 KRF93811.1 GL442330 EFN63568.1 CM000162 EDX02549.1 CH940651 EDW65554.1 CH916371 EDV92147.1 NNAY01002105 OXU22070.1 AJVK01034775
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000007266
+ More
UP000192223 UP000183832 UP000019118 UP000027135 UP000069940 UP000249989 UP000009046 UP000245037 UP000030742 UP000008820 UP000075920 UP000076408 UP000075900 UP000075880 UP000075886 UP000075901 UP000030765 UP000002320 UP000075902 UP000075840 UP000075885 UP000075903 UP000075884 UP000075882 UP000007062 UP000079169 UP000095301 UP000075883 UP000095300 UP000092444 UP000075881 UP000037069 UP000007755 UP000092443 UP000092460 UP000078200 UP000078492 UP000092445 UP000007798 UP000078540 UP000005203 UP000242457 UP000075809 UP000076407 UP000091820 UP000053097 UP000279307 UP000076502 UP000078541 UP000078542 UP000069272 UP000053825 UP000000803 UP000007819 UP000192221 UP000008711 UP000015103 UP000008237 UP000092553 UP000007801 UP000009192 UP000000311 UP000002282 UP000008792 UP000001070 UP000215335 UP000002358 UP000092462
UP000192223 UP000183832 UP000019118 UP000027135 UP000069940 UP000249989 UP000009046 UP000245037 UP000030742 UP000008820 UP000075920 UP000076408 UP000075900 UP000075880 UP000075886 UP000075901 UP000030765 UP000002320 UP000075902 UP000075840 UP000075885 UP000075903 UP000075884 UP000075882 UP000007062 UP000079169 UP000095301 UP000075883 UP000095300 UP000092444 UP000075881 UP000037069 UP000007755 UP000092443 UP000092460 UP000078200 UP000078492 UP000092445 UP000007798 UP000078540 UP000005203 UP000242457 UP000075809 UP000076407 UP000091820 UP000053097 UP000279307 UP000076502 UP000078541 UP000078542 UP000069272 UP000053825 UP000000803 UP000007819 UP000192221 UP000008711 UP000015103 UP000008237 UP000092553 UP000007801 UP000009192 UP000000311 UP000002282 UP000008792 UP000001070 UP000215335 UP000002358 UP000092462
PRIDE
Interpro
IPR036869
J_dom_sf
+ More
IPR001623 DnaJ_domain
IPR007829 TM2
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR036907 5'-Nucleotdase_C_sf
IPR006146 5'-Nucleotdase_CS
IPR033116 TRYPSIN_SER
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
IPR001623 DnaJ_domain
IPR007829 TM2
IPR006179 5_nucleotidase/apyrase
IPR029052 Metallo-depent_PP-like
IPR008334 5'-Nucleotdase_C
IPR004843 Calcineurin-like_PHP_ApaH
IPR036907 5'-Nucleotdase_C_sf
IPR006146 5'-Nucleotdase_CS
IPR033116 TRYPSIN_SER
IPR018114 TRYPSIN_HIS
IPR001254 Trypsin_dom
IPR009003 Peptidase_S1_PA
Gene 3D
ProteinModelPortal
H6VTQ8
A0A194PYF7
A0A194R027
A0A2A4JJX6
A0A212FM98
A0A2H1VN18
+ More
D6WIK7 A0A1W4WE44 A0A1Y1MMX0 A0A1J1IQG1 N6T5Z5 A0A067RPR7 A0A182GIN4 E0VZD7 A0A182GPW6 A0A2P8Z3X9 A0A336LMD3 A0A336M1U2 U4UG80 Q1HRR7 Q16UA2 A0A182WLK1 A0A182YKS5 A0A182RDM9 U5EZU4 A0A182J498 A0A182QAH7 A0A2M4CW11 A0A182ST93 A0A2M4BPM7 A0A2M3ZF21 A0A2M3YXG4 A0A2M4CW10 A0A1L8DTL0 A0A2M4A064 A0A2M3YXG5 A0A084WEL7 A0A2M3ZZ44 B0XGC5 A0A182TGZ2 A0A182I2K5 A0A182PCT1 A0A182V5G4 A0A182NK41 A0A182KQG8 A0A1Q3FDP8 Q7PTB4 A0A1S3D130 A0A1I8MEP3 A0A182LVJ1 A0A1I8Q1R3 A0A1B0G7E9 A0A182JP58 A0A0L0C1R2 F4WG03 A0A1A9XZK4 A0A1B0BYV7 A0A1A9VNL4 A0A195EGD5 A0A1A9ZUZ9 B4NPR9 A0A195BG49 W8CE63 A0A088A3K8 A0A2A3ESE4 A0A151XBM1 A0A0A1WPF6 A0A0K8UCV7 A0A182WX10 A0A1A9W119 A0A2H8TEW0 A0A026WSA5 A0A154P0K0 A0A069DSM4 A0A034VVG9 A0A195F0L6 A0A0V0G8R2 A0A0P4VWA9 A0A195BYS4 A0A182FH63 A0A2S2PCD9 A0A0L7QS91 Q9VX95 A0A2S2QN34 E9IN27 J9K8L7 A0A1W4VVB8 B3NWA2 T1I789 E2BVB1 A0A0M4FA27 B3N0B7 B4L230 E2ASK8 B4Q1X4 B4M1K0 B4JN54 A0A232EUN0 K7IN06 A0A1B0EZR6
D6WIK7 A0A1W4WE44 A0A1Y1MMX0 A0A1J1IQG1 N6T5Z5 A0A067RPR7 A0A182GIN4 E0VZD7 A0A182GPW6 A0A2P8Z3X9 A0A336LMD3 A0A336M1U2 U4UG80 Q1HRR7 Q16UA2 A0A182WLK1 A0A182YKS5 A0A182RDM9 U5EZU4 A0A182J498 A0A182QAH7 A0A2M4CW11 A0A182ST93 A0A2M4BPM7 A0A2M3ZF21 A0A2M3YXG4 A0A2M4CW10 A0A1L8DTL0 A0A2M4A064 A0A2M3YXG5 A0A084WEL7 A0A2M3ZZ44 B0XGC5 A0A182TGZ2 A0A182I2K5 A0A182PCT1 A0A182V5G4 A0A182NK41 A0A182KQG8 A0A1Q3FDP8 Q7PTB4 A0A1S3D130 A0A1I8MEP3 A0A182LVJ1 A0A1I8Q1R3 A0A1B0G7E9 A0A182JP58 A0A0L0C1R2 F4WG03 A0A1A9XZK4 A0A1B0BYV7 A0A1A9VNL4 A0A195EGD5 A0A1A9ZUZ9 B4NPR9 A0A195BG49 W8CE63 A0A088A3K8 A0A2A3ESE4 A0A151XBM1 A0A0A1WPF6 A0A0K8UCV7 A0A182WX10 A0A1A9W119 A0A2H8TEW0 A0A026WSA5 A0A154P0K0 A0A069DSM4 A0A034VVG9 A0A195F0L6 A0A0V0G8R2 A0A0P4VWA9 A0A195BYS4 A0A182FH63 A0A2S2PCD9 A0A0L7QS91 Q9VX95 A0A2S2QN34 E9IN27 J9K8L7 A0A1W4VVB8 B3NWA2 T1I789 E2BVB1 A0A0M4FA27 B3N0B7 B4L230 E2ASK8 B4Q1X4 B4M1K0 B4JN54 A0A232EUN0 K7IN06 A0A1B0EZR6
PDB
2CUG
E-value=1.43579e-07,
Score=131
Ontologies
GO
PANTHER
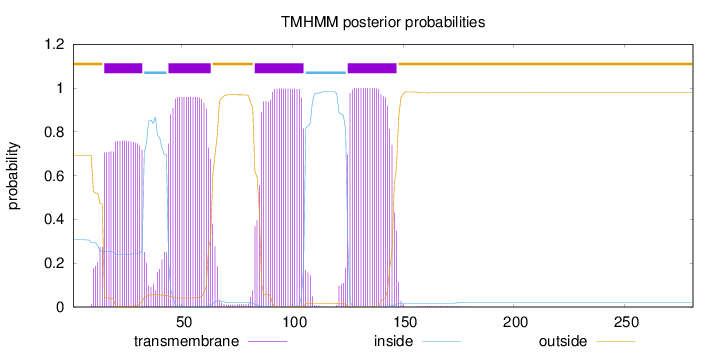
Topology
Length:
281
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
78.4129400000001
Exp number, first 60 AAs:
32.21606
Total prob of N-in:
0.30675
POSSIBLE N-term signal
sequence
outside
1 - 14
TMhelix
15 - 32
inside
33 - 43
TMhelix
44 - 63
outside
64 - 82
TMhelix
83 - 105
inside
106 - 124
TMhelix
125 - 147
outside
148 - 281
Population Genetic Test Statistics
Pi
17.729411
Theta
16.649943
Tajima's D
0.392899
CLR
0.538248
CSRT
0.486675666216689
Interpretation
Uncertain
