Pre Gene Modal
BGIBMGA000603
Annotation
PREDICTED:_JNK-interacting_protein_3_isoform_X2_[Bombyx_mori]
Full name
C-Jun-amino-terminal kinase-interacting protein 4
Alternative Name
JNK-associated leucine-zipper protein
JNK/SAPK-associated protein 2
Mitogen-activated protein kinase 8-interacting protein 4
Sperm-associated antigen 9
JNK/SAPK-associated protein 2
Mitogen-activated protein kinase 8-interacting protein 4
Sperm-associated antigen 9
Location in the cell
Nuclear Reliability : 3.454
Sequence
CDS
ATGAATTTCGACGACAGTGCGAGCAGCGGGTATGTTACCAATGAGACCATATACGGAACTCATGAAGATTCTCACGTAGTTATGTCAGAGAAGGTACAATCTCTTGCTGGAAGTATCTACCAAGAGTTCGAACGTATGATAGCGCGATATGATGAGGATGTAGTAAAAACATTGATGCCACTTCTAGTGAATGTATTAGAGTGCTTGGATTCAGCCTATCAGACGAATCAAGAGCACGAGGTGGAACTAGAGCTATTGCGAGAAGACAACGAGCAGTTAGTTACGCAGTATGAACGCGAGAAGGCAGCCAGGAAGTACGCTGAACAGAAGTTACTGGAAGCTGAGGATCATTATGAAGGAGAAAGAAAAGACTTGACTGGCCGTTTAGAAGCACTGGATAGTATCGTGAGAATGTTAGAACTTAAACACAAGAACTCCTTGGATCATGCTAGTAGACTCGAAGAAAGAGAAAATGAATTGAAAAAAGAATATGCAAAGTTGCATGAAAGGTACACCGAGCTATTTAAAACTCACATGGACTATATGGAAAGAACAAAGCTACTTCTAAGCACAGCAAGTGATAGAAGCGAAGTTCGAGGCCGACTGGGACTCAATCCTGTTGGAAGGTCTAGTGGTCCGATATCGTTTGGTTTCGCATCTCTAGAAAGTTCTATGAACGCGATAGAACAGACCGAAAGTATTCCTGGTAGTCCTGTCAGCTTGTCGTCGTCACCGCCATCGCCGCCCATCGGAGCCGAACTAGCGGCCGTGAGGACGGTCGAAAGAGCTCACCAGACAGATGCCATAACACAACAAAGTCAGGCAACGTCGCCTGTCGCTCTCCCGAATTCAAACATCGGAAAATCTCAGACTAAGAGCGAAAAACGAAGTGGTAACACTTTGTATCAGGAGTTGGCTTTCCATGACACCGATGCGAGCATTGCCGAATCACAAGGCGATGAAGGTGCTGACATCACAGGTAGCTGGGTGCATCCTGGAGAATATGCGTCTTCTGGCATGGGGAAAGAAGTCGAAAACTTGATACTGGAGAACAACGAATTATTGGCCACGAAGAACGCTTTGAACGTTGTCAAGGACGACCTTATAGTGAAGGTGGACGAGTTGACCGGCGAACAGGAGATGCTTCGAGAAGAAGTGGCCAATCTAAGTTCGGCGCGAGAGAAACTTAGAGATCGGGTGACTCAATTGGAGGATGAATTGCGACGTTTGAAGGAGGCGATGAGCAACAACGCCGGCGGTGGGGACAACGAGGAGGAAGCCGACGTCCCGTTGGCGCAGAGACGCCGCTTCACGCGCGTCGAGATGGCACGTGTCCTCATGGAACGTAACCAATATAAGGAACGATTAATGGAGCTTCAAGATGCCGTTCGCTGGACCGAGATGGTTCGTGCCAGTCGCGTGGACAACTCCATCGACAAGAAGAGCAAGCAGAGCATATGGACTTTTTTTAGTAAACTATTCAGTTCGAACGATCGTCCACAAAGACCGCTTTCCACTCCTCATCTAAGAGCCGTGGCCGCCTCACAGCCTACGATGAGACATTCACAAACCCAAGGCGACTTCTTGGAGGCGCAGCTTTCCGACCACCACCAGAAGCGACACGAGAGAACCGAACAATTCCGACAGGTGCGGGTACATGTGCGTAAGGAAGACGGGCGCACGCACGCTTACGGTTGGAGTCTCCCGGGGAAGAACAGCCAGGGACAGAAGGGACCGAGCCCCTCAGCGTCCCTGTCGTCGGGGGGAGTTCCGGTACCAGTTCCGGTTTTTTGTCGCCCGATAGCGGAGAGTGAGCCGCGTATGACTCTGCTGTGTGCGGCCGGCGTAAACCTGAACGGAGGTAGGACTCCGGATGGAGGCTGCATGATTGGTGAGAGCGTTTTCTACGCAAATGAAGACGATAAGACGGAGAACAATATGTCCTTAGCCTACGAGATGAGCAAAGCGCAGTCAGCGCATTCTCCGGAGGTCGAGCAGATCAGCAGACAGCTGCAGTCAACTCAGAGCGTGGAGTACACTGAAAAGAATCTGTCCTCGTTGGTTTGGATATGTGCTAGTACACAGGCTAAAGGAGTGGTTATGATCATCGATGCGAACAATCCGGCGGACGTGATCGAATCCTTTTCAGTGAGCGATAAGCATATTCTCTGTGTAGCTTCAGTGCCTGGTGCCACTTCAGAAGATTATCAAGATTACGAAGAACGAATGTCTGGGGACACCGAAACGAAGCGTCATTCGTCGTTGTTATGCGTGGGAAAGAGTGAGTCTGTGGAGAGCCAACCTAATGGCCCCGTCGAGGACACGAACACGGAGAACACTGAAAACGAAACTGTAATTGGAACTACTAAATTGATAAAGGCAACCCTTCTAACTCCACCGAGTACTCCCGTGAAGGAGCCAGAAGAACCAGCTGCTGATAATGACGAGAATCCCAAAGAATCGATGGTCAAGAATGTTGAAGACGAAGCAGAGAACGACGGTCATGACAGTCCGCCACTAAGCTCCACTCCTAATAATGTAACGGAGCCATCGGTCAGTTCACCGGAGCCCGGATTGGACGCGTTCGCACAGGAACAATCGAACGTCGAGAACCGCGGCGATGTACCGGACCGAAAGATGTCCACCGTGATGGCGACGATGTGGCTAGGAACCAAAAACGGAAACCTATACGTACATTCCGCAATGGGAAACTACAGCAAGTGCTTGGCAAGGATCAAACTGAACGATGCTATATTGGCCATAGTGTCGTGTGCTTCTCGTTGCGTGGTCGCTCTCGCTGATGGAACGGTAGCTATATTTGCCCGACTTGCAGATGGACAGTGGGATTTCACTCAGTATTGGCTCATGACTCTAGGCGATTCTAAATGTTCAGTGCGATGTCTCAGTGCGGTCGGCTGTACGGTTTGGTGCGGTTACCGCAACAAGGTCCAGGTGATCGATCCTCGTACGAGACAACTACTTCACACCCTCGAAGCTCATCCTAGACAAGAGAGTCAAGTGCGACAGATGGCGTGTCACGGCGATGGAGTATGGGTATCGATAAAAATGGACTCCACCCTCGGTCTGTACCACGCGCACACATACAAGAACTTGTGCAACGTCGACATCGAACCGTACGTGAGCAAAATGCTCGGAACGGGCAAACTTGGATTCTCTCTGGTCAGGATCACGGCGCTTCTAATCAGTTCCGGGCGTCTTTGGATCGGAACCAGCAATGGGGTTGTTATATCCGTGCCACTGTCCGAAGCTAATCGAGATCCGATTCATGCAACATTAAATCGAACGAGATCCGCGAGTCCGACTGGGGTTATACCCTGGTGCTCGATGGCTCACGCCCAGCTCAGCTTCCACGGTCACCGGGACGCGGTCAAGTTCTTCGTGGCGGTACCGGGGGCCGCTAGTTCGTCAGCCAGAACACCGGAGTCGCCCCAGCAAGGTTCGAACCAGCCGCCGATGCTGGTCATATCCGGAGGCGAGGGATATATTGACTTTAGAATAGCCGACTCGGAGATGGAGGACAGCGTGGTGGTAGCCGAAGATGGTTCCTCGAGTGGCCACAGCTCGTTAATTGAGAGGGCCTCTAAGAGTCACTTGATAGTGTGGCAAGTAGCGACGGCCTAA
Protein
MNFDDSASSGYVTNETIYGTHEDSHVVMSEKVQSLAGSIYQEFERMIARYDEDVVKTLMPLLVNVLECLDSAYQTNQEHEVELELLREDNEQLVTQYEREKAARKYAEQKLLEAEDHYEGERKDLTGRLEALDSIVRMLELKHKNSLDHASRLEERENELKKEYAKLHERYTELFKTHMDYMERTKLLLSTASDRSEVRGRLGLNPVGRSSGPISFGFASLESSMNAIEQTESIPGSPVSLSSSPPSPPIGAELAAVRTVERAHQTDAITQQSQATSPVALPNSNIGKSQTKSEKRSGNTLYQELAFHDTDASIAESQGDEGADITGSWVHPGEYASSGMGKEVENLILENNELLATKNALNVVKDDLIVKVDELTGEQEMLREEVANLSSAREKLRDRVTQLEDELRRLKEAMSNNAGGGDNEEEADVPLAQRRRFTRVEMARVLMERNQYKERLMELQDAVRWTEMVRASRVDNSIDKKSKQSIWTFFSKLFSSNDRPQRPLSTPHLRAVAASQPTMRHSQTQGDFLEAQLSDHHQKRHERTEQFRQVRVHVRKEDGRTHAYGWSLPGKNSQGQKGPSPSASLSSGGVPVPVPVFCRPIAESEPRMTLLCAAGVNLNGGRTPDGGCMIGESVFYANEDDKTENNMSLAYEMSKAQSAHSPEVEQISRQLQSTQSVEYTEKNLSSLVWICASTQAKGVVMIIDANNPADVIESFSVSDKHILCVASVPGATSEDYQDYEERMSGDTETKRHSSLLCVGKSESVESQPNGPVEDTNTENTENETVIGTTKLIKATLLTPPSTPVKEPEEPAADNDENPKESMVKNVEDEAENDGHDSPPLSSTPNNVTEPSVSSPEPGLDAFAQEQSNVENRGDVPDRKMSTVMATMWLGTKNGNLYVHSAMGNYSKCLARIKLNDAILAIVSCASRCVVALADGTVAIFARLADGQWDFTQYWLMTLGDSKCSVRCLSAVGCTVWCGYRNKVQVIDPRTRQLLHTLEAHPRQESQVRQMACHGDGVWVSIKMDSTLGLYHAHTYKNLCNVDIEPYVSKMLGTGKLGFSLVRITALLISSGRLWIGTSNGVVISVPLSEANRDPIHATLNRTRSASPTGVIPWCSMAHAQLSFHGHRDAVKFFVAVPGAASSSARTPESPQQGSNQPPMLVISGGEGYIDFRIADSEMEDSVVVAEDGSSSGHSSLIERASKSHLIVWQVATA
Summary
Description
The JNK-interacting protein (JIP) group of scaffold proteins selectively mediates JNK signaling by aggregating specific components of the MAPK cascade to form a functional JNK signaling module.
Subunit
Homodimer. The homodimer interacts with ARF6, forming a heterotetramer (By similarity). Homooligomer. Interacts with MAX, MAPK8, MAPK14, MAPK10, MAPK14, MAP3K3, MYC, KNS2, and MAP2K4. Interaction with KNS2 is important in the formation of ternary complex with MAPK8.
Similarity
Belongs to the JIP scaffold family.
Keywords
Acetylation
Alternative splicing
Coiled coil
Complete proteome
Cytoplasm
Phosphoprotein
Reference proteome
Feature
chain C-Jun-amino-terminal kinase-interacting protein 4
splice variant In isoform 6.
splice variant In isoform 6.
Uniprot
H9ITM3
A0A2H1VN22
A0A194Q4Y3
A0A212FMC1
V5GTY7
V5GI32
+ More
A0A154PDH8 A0A2A3EP43 A0A0N0U630 A0A195E439 A0A1B6HN80 A0A0L7R9H8 A0A1B6DJ19 A0A195BU43 A0A310SKA3 A0A158P2A9 A0A1B6IUX2 A0A232EYH2 A0A1J1I612 E2AJX5 Q16LZ1 A0A1B6FKZ4 U4U4V0 N6TG04 A0A182H5F5 A0A1B6DA52 A0A1B6E7J4 Q16LZ0 A0A0Q9WUY8 A0A023F2S2 A0A336LP60 A0A0Q9WXB8 A0A0Q9WVQ7 A0A0Q9XMH0 A0A1Q3G3V7 B4LHS0 A0A0C9RUK7 A0A069DXF2 A0A0Q9WJT5 A0A0Q9XJV9 A0A0Q9XEM4 A0A1A9XHV8 B4KUP2 A0A0Q9WKM9 A0A0Q9XPR3 A0A0V0G5K6 A0A0Q9WTS3 A0A1B6MMC8 A0A0Q9WVP6 A0A0Q9XC56 A0A1Q3G3Q0 A0A1B6MDW4 A0A1Q3G3R5 A0A0Q9XCA3 A0A0Q9XC34 A0A1Q3G3Q1 U4TXU9 N6UDF1 A0A0Q9XC33 A0A1A9WCE0 T1IY26 A0A2R5LMW2 A0A131XZH8 V5I520 A0A1L8DLI1 A0A1L8DLF4 A0A224X641 A0A087U4F5 A0A1S3IWE9 A0A1S3HFK4 A0A1S3HIM1 A0A1B6CAD9 R7V594 A0A1S2ZWB5 A0A1S2ZWB6 Q58A65-3 Q58A65-2 Q58A65 A0A1S2ZWC4 Q58A65-5 E9PUD1 A0A287AW88 A0A0R4J196 A0A1S2ZWC3 Q58A65-6 A0A337SBI1
A0A154PDH8 A0A2A3EP43 A0A0N0U630 A0A195E439 A0A1B6HN80 A0A0L7R9H8 A0A1B6DJ19 A0A195BU43 A0A310SKA3 A0A158P2A9 A0A1B6IUX2 A0A232EYH2 A0A1J1I612 E2AJX5 Q16LZ1 A0A1B6FKZ4 U4U4V0 N6TG04 A0A182H5F5 A0A1B6DA52 A0A1B6E7J4 Q16LZ0 A0A0Q9WUY8 A0A023F2S2 A0A336LP60 A0A0Q9WXB8 A0A0Q9WVQ7 A0A0Q9XMH0 A0A1Q3G3V7 B4LHS0 A0A0C9RUK7 A0A069DXF2 A0A0Q9WJT5 A0A0Q9XJV9 A0A0Q9XEM4 A0A1A9XHV8 B4KUP2 A0A0Q9WKM9 A0A0Q9XPR3 A0A0V0G5K6 A0A0Q9WTS3 A0A1B6MMC8 A0A0Q9WVP6 A0A0Q9XC56 A0A1Q3G3Q0 A0A1B6MDW4 A0A1Q3G3R5 A0A0Q9XCA3 A0A0Q9XC34 A0A1Q3G3Q1 U4TXU9 N6UDF1 A0A0Q9XC33 A0A1A9WCE0 T1IY26 A0A2R5LMW2 A0A131XZH8 V5I520 A0A1L8DLI1 A0A1L8DLF4 A0A224X641 A0A087U4F5 A0A1S3IWE9 A0A1S3HFK4 A0A1S3HIM1 A0A1B6CAD9 R7V594 A0A1S2ZWB5 A0A1S2ZWB6 Q58A65-3 Q58A65-2 Q58A65 A0A1S2ZWC4 Q58A65-5 E9PUD1 A0A287AW88 A0A0R4J196 A0A1S2ZWC3 Q58A65-6 A0A337SBI1
Pubmed
EMBL
BABH01006694
ODYU01003434
SOQ42196.1
KQ459463
KPJ00598.1
AGBW02007654
+ More
OWR54877.1 GALX01004763 JAB63703.1 GALX01004762 JAB63704.1 KQ434869 KZC09250.1 KZ288202 PBC33487.1 KQ435742 KOX76702.1 KQ979657 KYN19950.1 GECU01031574 JAS76132.1 KQ414625 KOC67530.1 GEDC01011646 JAS25652.1 KQ976408 KYM91134.1 KQ762031 OAD56376.1 ADTU01007051 GECU01016967 JAS90739.1 NNAY01001595 OXU23482.1 CVRI01000042 CRK95704.1 GL440100 EFN66291.1 CH477886 EAT35356.1 GECZ01018914 JAS50855.1 KB631730 ERL85636.1 APGK01039256 KB740969 ENN76688.1 JXUM01004247 KQ560171 KXJ84024.1 GEDC01014798 JAS22500.1 GEDC01003395 JAS33903.1 EAT35357.1 CH940647 KRF85039.1 GBBI01003224 JAC15488.1 UFQS01000097 UFQT01000097 SSW99416.1 SSX19796.1 KRF85040.1 KRF85038.1 CH933809 KRG06110.1 GFDL01000596 JAV34449.1 EDW70645.1 GBYB01011271 JAG81038.1 GBGD01000146 JAC88743.1 KRF85037.1 KRG06106.1 KRG06105.1 EDW18270.1 KRF85042.1 KRG06104.1 GECL01002795 JAP03329.1 KRF85041.1 GEBQ01002879 JAT37098.1 KRF85043.1 KRF85044.1 KRG06107.1 GFDL01000610 JAV34435.1 GEBQ01005850 JAT34127.1 GFDL01000603 JAV34442.1 KRG06108.1 KRG06103.1 KRG06111.1 GFDL01000613 JAV34432.1 ERL85637.1 ENN76687.1 KRG06112.1 JH431669 GGLE01006703 MBY10829.1 GEFM01003377 JAP72419.1 GANP01000740 JAB83728.1 GFDF01006782 JAV07302.1 GFDF01006781 JAV07303.1 GFTR01008479 JAW07947.1 KK118105 KFM72244.1 GEDC01026877 JAS10421.1 AMQN01005076 KB295063 ELU13632.1 AF327451 AY823270 AB047782 AK172961 AK147431 AK147537 AL662838 BC060100 BC094670 AEMK02000080 AANG04003358
OWR54877.1 GALX01004763 JAB63703.1 GALX01004762 JAB63704.1 KQ434869 KZC09250.1 KZ288202 PBC33487.1 KQ435742 KOX76702.1 KQ979657 KYN19950.1 GECU01031574 JAS76132.1 KQ414625 KOC67530.1 GEDC01011646 JAS25652.1 KQ976408 KYM91134.1 KQ762031 OAD56376.1 ADTU01007051 GECU01016967 JAS90739.1 NNAY01001595 OXU23482.1 CVRI01000042 CRK95704.1 GL440100 EFN66291.1 CH477886 EAT35356.1 GECZ01018914 JAS50855.1 KB631730 ERL85636.1 APGK01039256 KB740969 ENN76688.1 JXUM01004247 KQ560171 KXJ84024.1 GEDC01014798 JAS22500.1 GEDC01003395 JAS33903.1 EAT35357.1 CH940647 KRF85039.1 GBBI01003224 JAC15488.1 UFQS01000097 UFQT01000097 SSW99416.1 SSX19796.1 KRF85040.1 KRF85038.1 CH933809 KRG06110.1 GFDL01000596 JAV34449.1 EDW70645.1 GBYB01011271 JAG81038.1 GBGD01000146 JAC88743.1 KRF85037.1 KRG06106.1 KRG06105.1 EDW18270.1 KRF85042.1 KRG06104.1 GECL01002795 JAP03329.1 KRF85041.1 GEBQ01002879 JAT37098.1 KRF85043.1 KRF85044.1 KRG06107.1 GFDL01000610 JAV34435.1 GEBQ01005850 JAT34127.1 GFDL01000603 JAV34442.1 KRG06108.1 KRG06103.1 KRG06111.1 GFDL01000613 JAV34432.1 ERL85637.1 ENN76687.1 KRG06112.1 JH431669 GGLE01006703 MBY10829.1 GEFM01003377 JAP72419.1 GANP01000740 JAB83728.1 GFDF01006782 JAV07302.1 GFDF01006781 JAV07303.1 GFTR01008479 JAW07947.1 KK118105 KFM72244.1 GEDC01026877 JAS10421.1 AMQN01005076 KB295063 ELU13632.1 AF327451 AY823270 AB047782 AK172961 AK147431 AK147537 AL662838 BC060100 BC094670 AEMK02000080 AANG04003358
Proteomes
UP000005204
UP000053268
UP000007151
UP000076502
UP000242457
UP000053105
+ More
UP000078492 UP000053825 UP000078540 UP000005205 UP000215335 UP000183832 UP000000311 UP000008820 UP000030742 UP000019118 UP000069940 UP000249989 UP000008792 UP000009192 UP000092443 UP000091820 UP000054359 UP000085678 UP000014760 UP000079721 UP000000589 UP000008227 UP000011712
UP000078492 UP000053825 UP000078540 UP000005205 UP000215335 UP000183832 UP000000311 UP000008820 UP000030742 UP000019118 UP000069940 UP000249989 UP000008792 UP000009192 UP000092443 UP000091820 UP000054359 UP000085678 UP000014760 UP000079721 UP000000589 UP000008227 UP000011712
PRIDE
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR032486 JIP_LZII
IPR034744 RH2
IPR034743 RH1
IPR039911 JIP3/JIP4
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019143 JNK/Rab-associated_protein-1_N
IPR011047 Quinoprotein_ADH-like_supfam
IPR005475 Transketolase-like_Pyr-bd
IPR009014 Transketo_C/PFOR_II
IPR029061 THDP-binding
IPR033248 Transketolase_C
IPR011044 Quino_amine_DH_bsu
IPR002119 Histone_H2A
IPR009072 Histone-fold
IPR032486 JIP_LZII
IPR034744 RH2
IPR034743 RH1
IPR039911 JIP3/JIP4
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR019143 JNK/Rab-associated_protein-1_N
IPR011047 Quinoprotein_ADH-like_supfam
IPR005475 Transketolase-like_Pyr-bd
IPR009014 Transketo_C/PFOR_II
IPR029061 THDP-binding
IPR033248 Transketolase_C
IPR011044 Quino_amine_DH_bsu
IPR002119 Histone_H2A
IPR009072 Histone-fold
SUPFAM
Gene 3D
ProteinModelPortal
H9ITM3
A0A2H1VN22
A0A194Q4Y3
A0A212FMC1
V5GTY7
V5GI32
+ More
A0A154PDH8 A0A2A3EP43 A0A0N0U630 A0A195E439 A0A1B6HN80 A0A0L7R9H8 A0A1B6DJ19 A0A195BU43 A0A310SKA3 A0A158P2A9 A0A1B6IUX2 A0A232EYH2 A0A1J1I612 E2AJX5 Q16LZ1 A0A1B6FKZ4 U4U4V0 N6TG04 A0A182H5F5 A0A1B6DA52 A0A1B6E7J4 Q16LZ0 A0A0Q9WUY8 A0A023F2S2 A0A336LP60 A0A0Q9WXB8 A0A0Q9WVQ7 A0A0Q9XMH0 A0A1Q3G3V7 B4LHS0 A0A0C9RUK7 A0A069DXF2 A0A0Q9WJT5 A0A0Q9XJV9 A0A0Q9XEM4 A0A1A9XHV8 B4KUP2 A0A0Q9WKM9 A0A0Q9XPR3 A0A0V0G5K6 A0A0Q9WTS3 A0A1B6MMC8 A0A0Q9WVP6 A0A0Q9XC56 A0A1Q3G3Q0 A0A1B6MDW4 A0A1Q3G3R5 A0A0Q9XCA3 A0A0Q9XC34 A0A1Q3G3Q1 U4TXU9 N6UDF1 A0A0Q9XC33 A0A1A9WCE0 T1IY26 A0A2R5LMW2 A0A131XZH8 V5I520 A0A1L8DLI1 A0A1L8DLF4 A0A224X641 A0A087U4F5 A0A1S3IWE9 A0A1S3HFK4 A0A1S3HIM1 A0A1B6CAD9 R7V594 A0A1S2ZWB5 A0A1S2ZWB6 Q58A65-3 Q58A65-2 Q58A65 A0A1S2ZWC4 Q58A65-5 E9PUD1 A0A287AW88 A0A0R4J196 A0A1S2ZWC3 Q58A65-6 A0A337SBI1
A0A154PDH8 A0A2A3EP43 A0A0N0U630 A0A195E439 A0A1B6HN80 A0A0L7R9H8 A0A1B6DJ19 A0A195BU43 A0A310SKA3 A0A158P2A9 A0A1B6IUX2 A0A232EYH2 A0A1J1I612 E2AJX5 Q16LZ1 A0A1B6FKZ4 U4U4V0 N6TG04 A0A182H5F5 A0A1B6DA52 A0A1B6E7J4 Q16LZ0 A0A0Q9WUY8 A0A023F2S2 A0A336LP60 A0A0Q9WXB8 A0A0Q9WVQ7 A0A0Q9XMH0 A0A1Q3G3V7 B4LHS0 A0A0C9RUK7 A0A069DXF2 A0A0Q9WJT5 A0A0Q9XJV9 A0A0Q9XEM4 A0A1A9XHV8 B4KUP2 A0A0Q9WKM9 A0A0Q9XPR3 A0A0V0G5K6 A0A0Q9WTS3 A0A1B6MMC8 A0A0Q9WVP6 A0A0Q9XC56 A0A1Q3G3Q0 A0A1B6MDW4 A0A1Q3G3R5 A0A0Q9XCA3 A0A0Q9XC34 A0A1Q3G3Q1 U4TXU9 N6UDF1 A0A0Q9XC33 A0A1A9WCE0 T1IY26 A0A2R5LMW2 A0A131XZH8 V5I520 A0A1L8DLI1 A0A1L8DLF4 A0A224X641 A0A087U4F5 A0A1S3IWE9 A0A1S3HFK4 A0A1S3HIM1 A0A1B6CAD9 R7V594 A0A1S2ZWB5 A0A1S2ZWB6 Q58A65-3 Q58A65-2 Q58A65 A0A1S2ZWC4 Q58A65-5 E9PUD1 A0A287AW88 A0A0R4J196 A0A1S2ZWC3 Q58A65-6 A0A337SBI1
PDB
4PXJ
E-value=1.66662e-09,
Score=155
Ontologies
GO
GO:0005078
GO:0003824
GO:0016301
GO:0030140
GO:0008088
GO:0043005
GO:0019894
GO:0016192
GO:0000786
GO:0003677
GO:0005634
GO:0046982
GO:0005737
GO:0000187
GO:0051260
GO:0005829
GO:0042147
GO:0048273
GO:0005815
GO:0008432
GO:0007257
GO:0030159
GO:0090074
GO:0043410
GO:0030335
GO:0045666
GO:0048471
GO:0051146
GO:0016021
GO:0005515
GO:0003676
GO:0000413
GO:0016020
GO:0043565
GO:0043401
GO:0003707
GO:0007169
PANTHER
Topology
Subcellular location
Cytoplasm
Perinuclear region
Perinuclear region
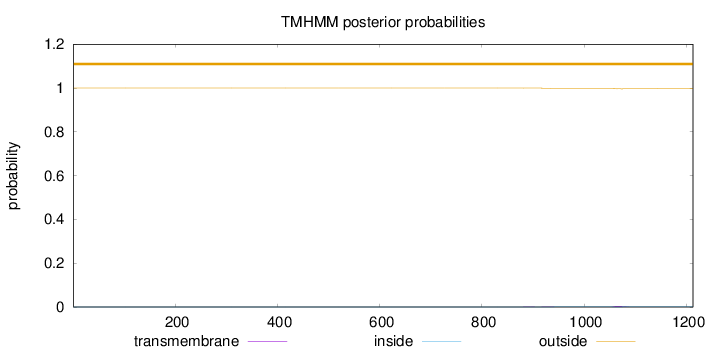
Length:
1213
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0955000000000001
Exp number, first 60 AAs:
7e-05
Total prob of N-in:
0.00001
outside
1 - 1213
Population Genetic Test Statistics
Pi
23.039178
Theta
20.859029
Tajima's D
-1.593878
CLR
0.022806
CSRT
0.0485975701214939
Interpretation
Uncertain
